Background
We have longitudinal data from 10 measurements.
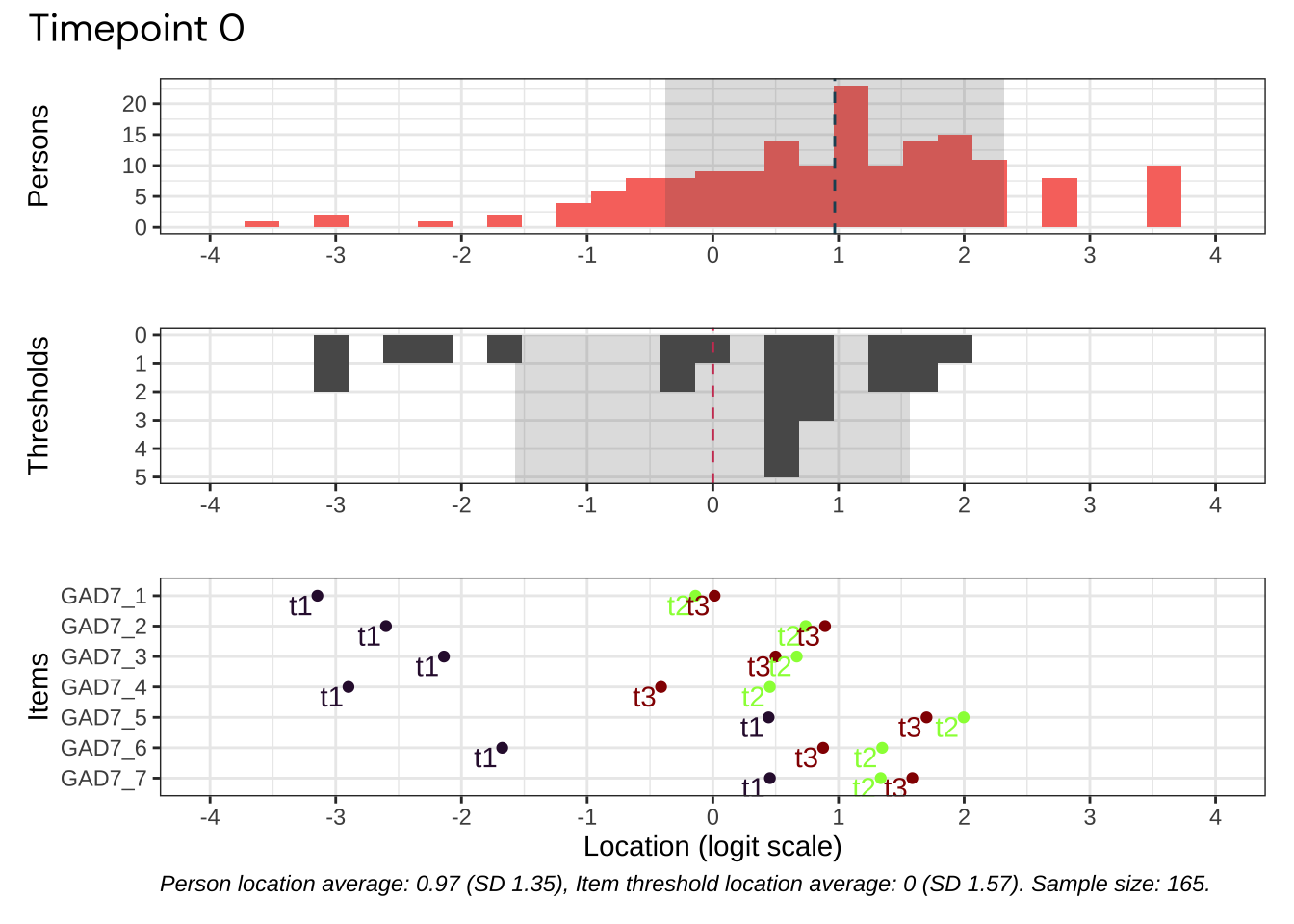
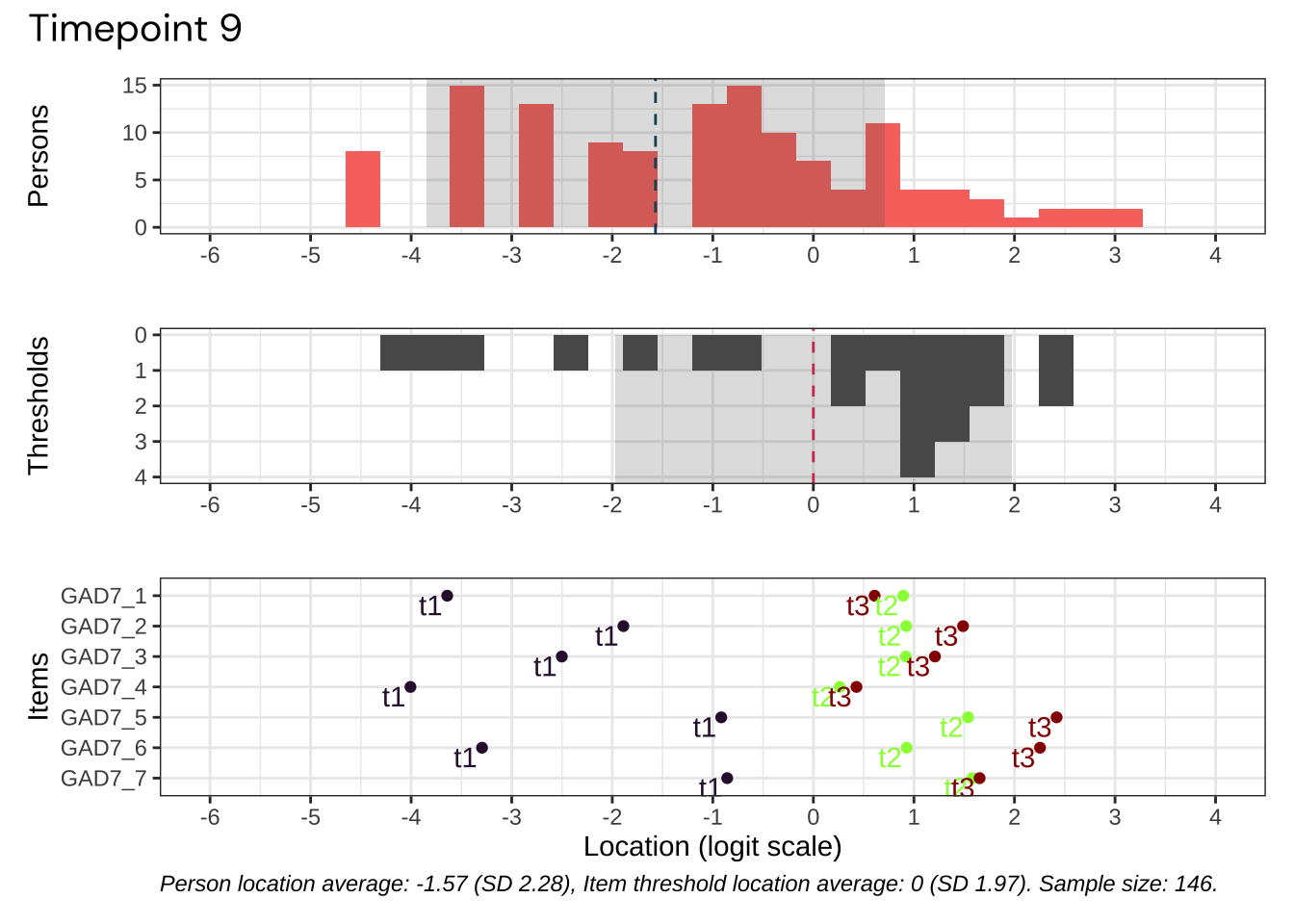
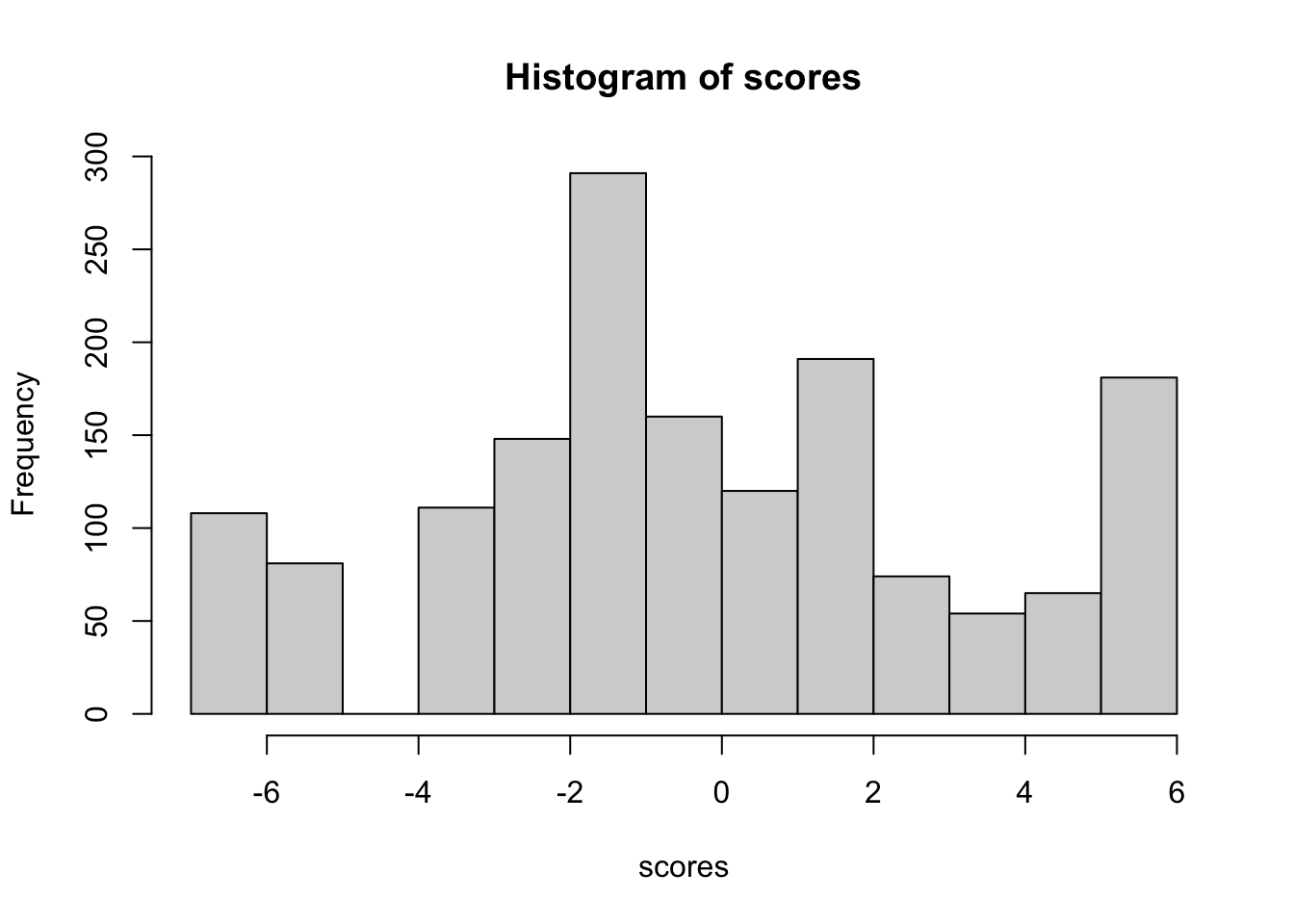
We’ll select the datapoint with best targeting for our psychometric analysis.
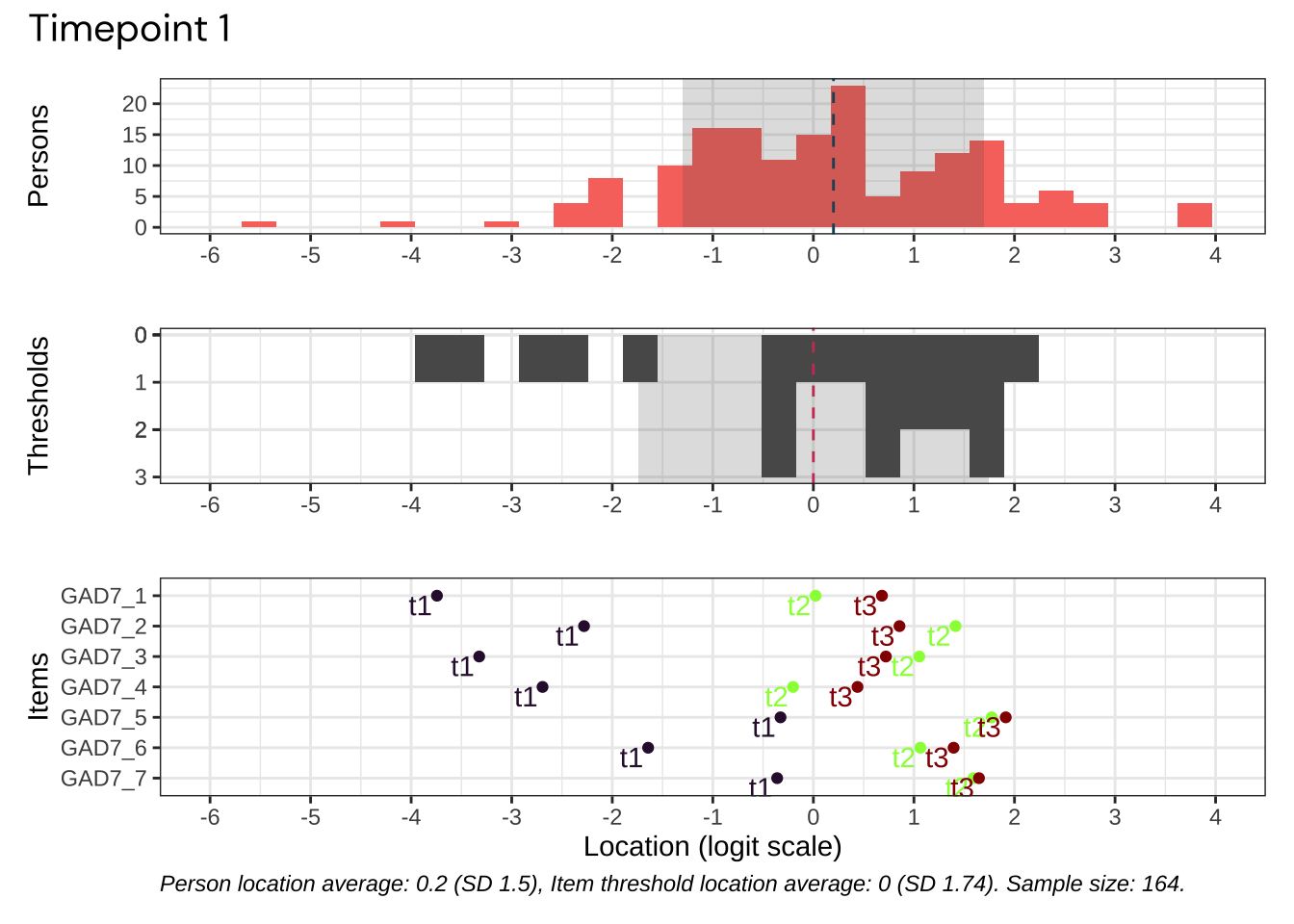
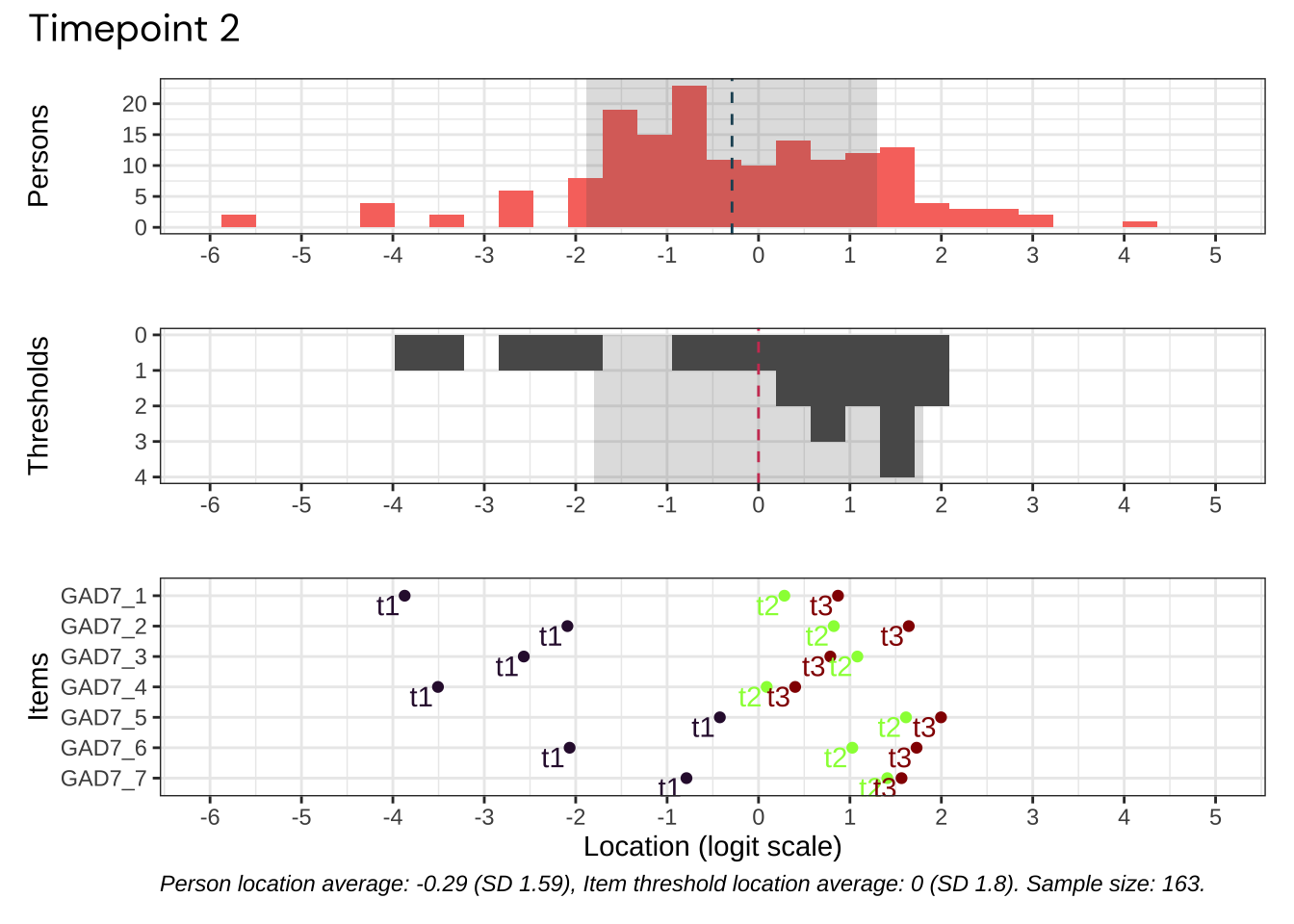
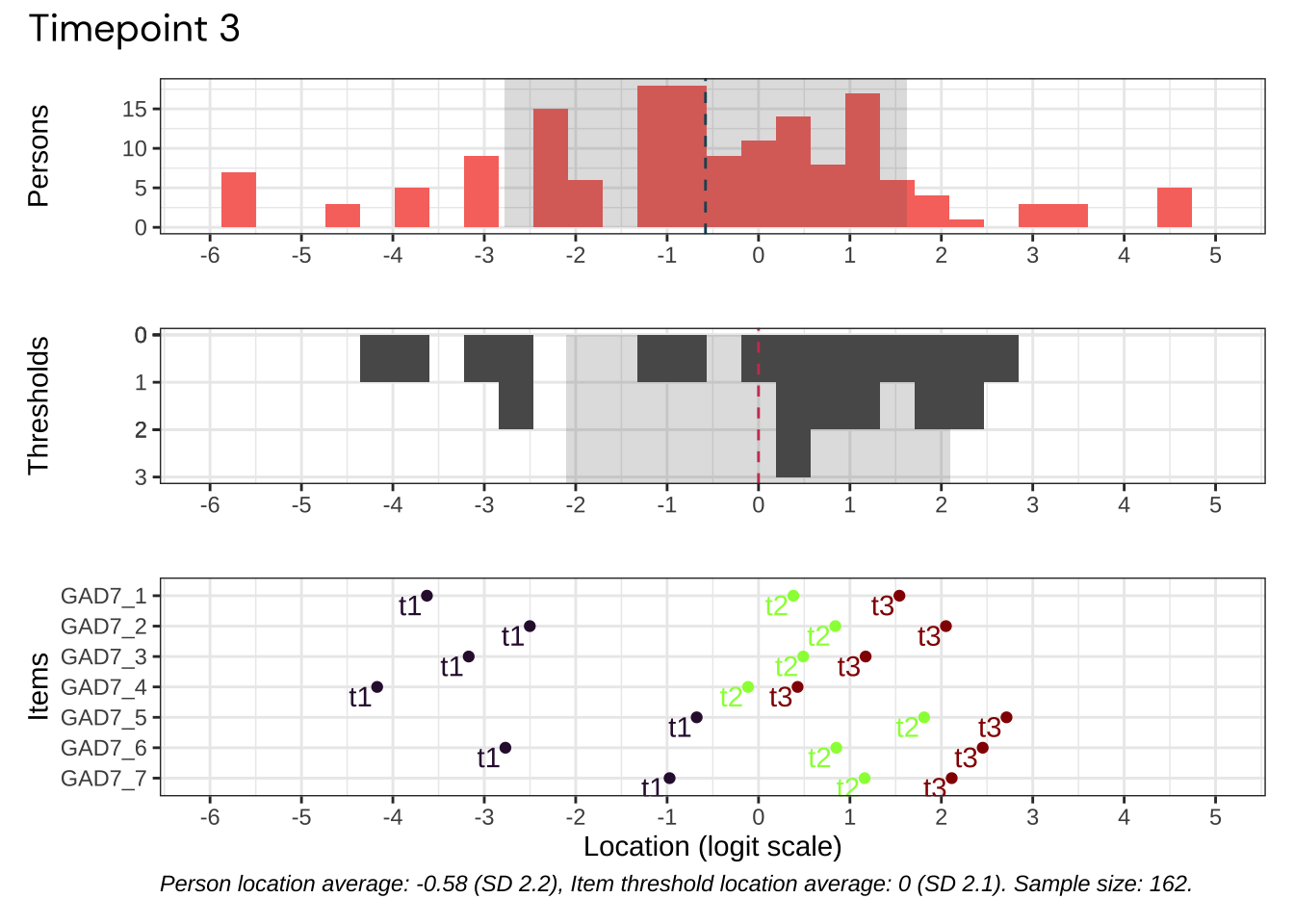
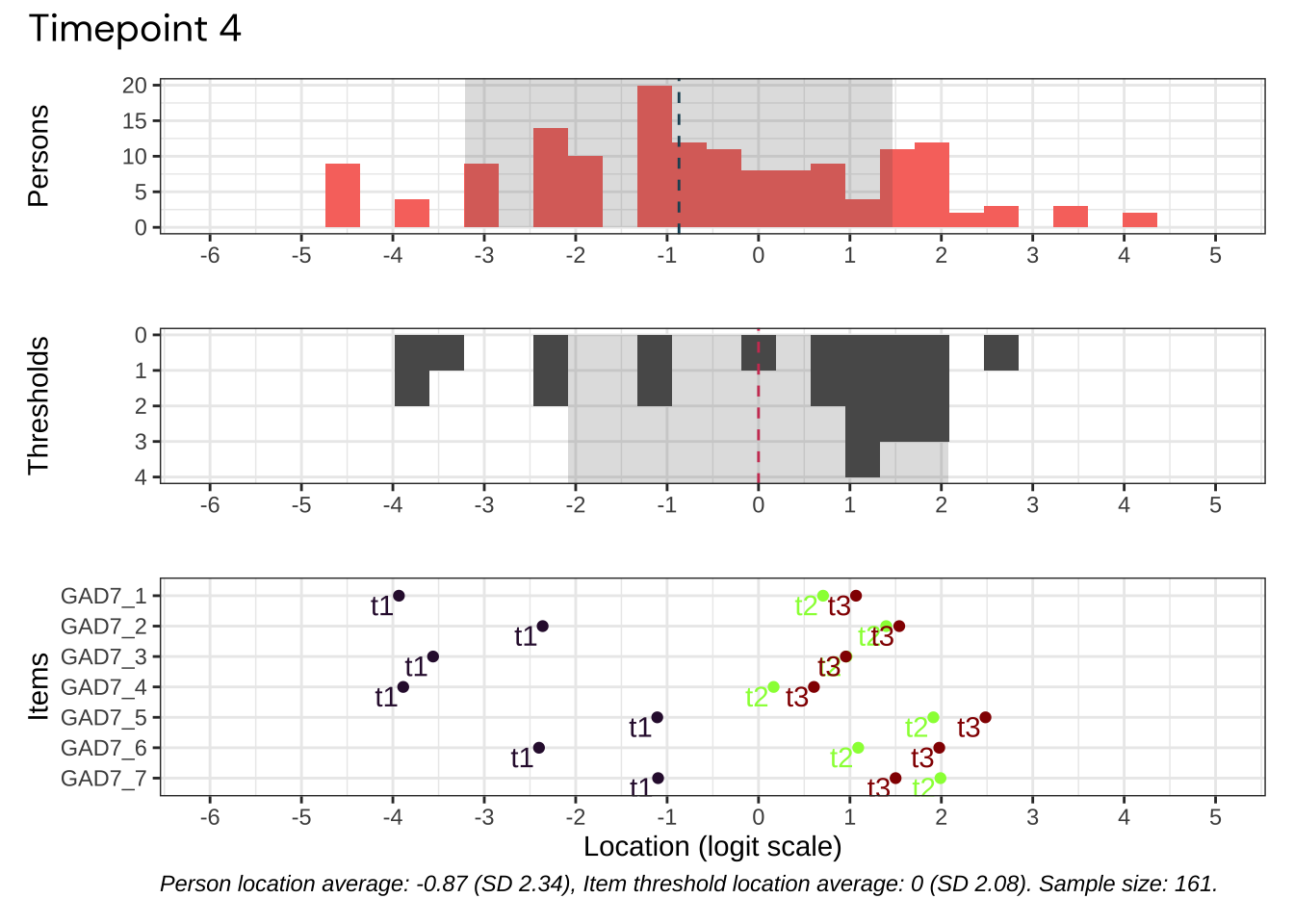
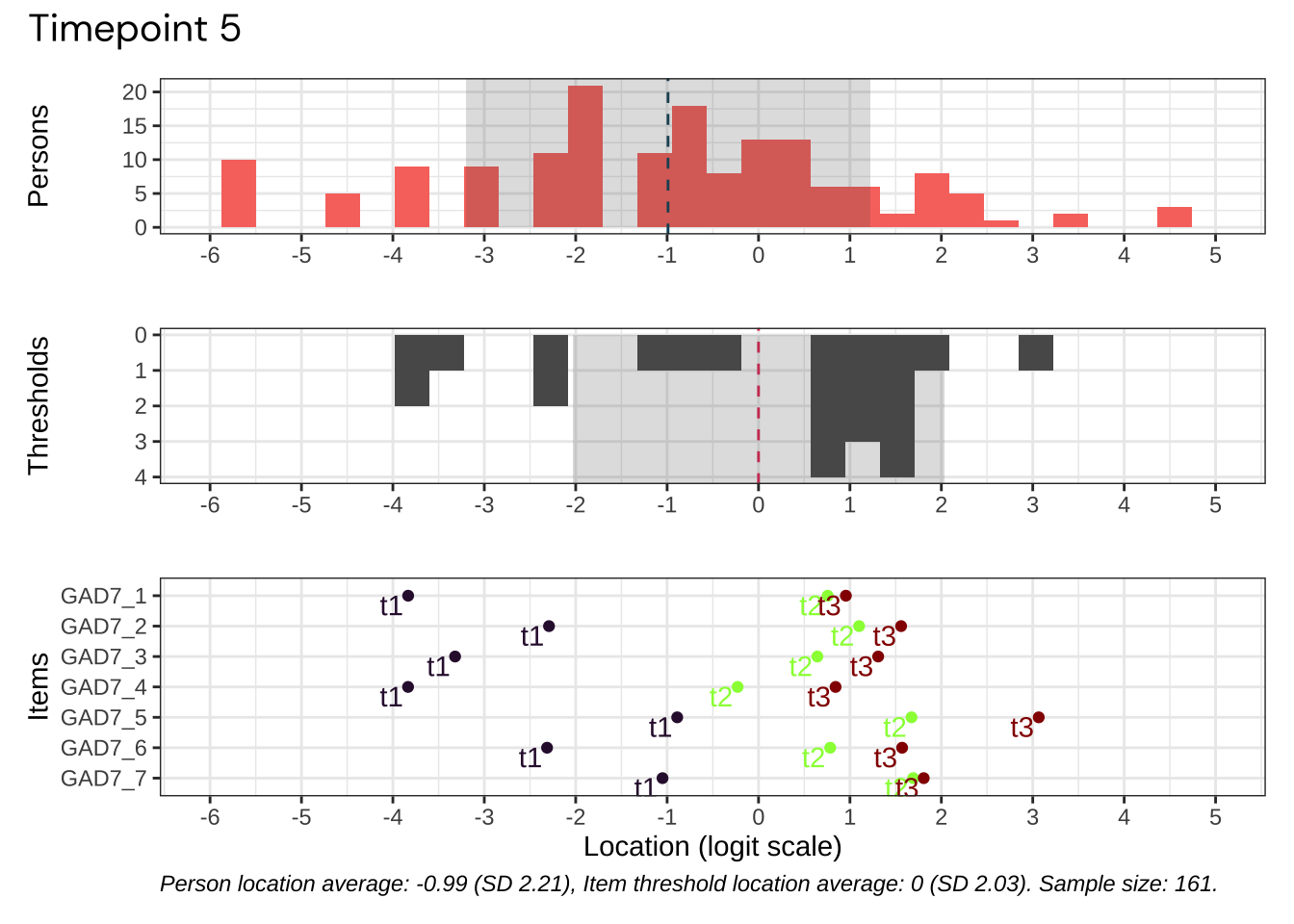
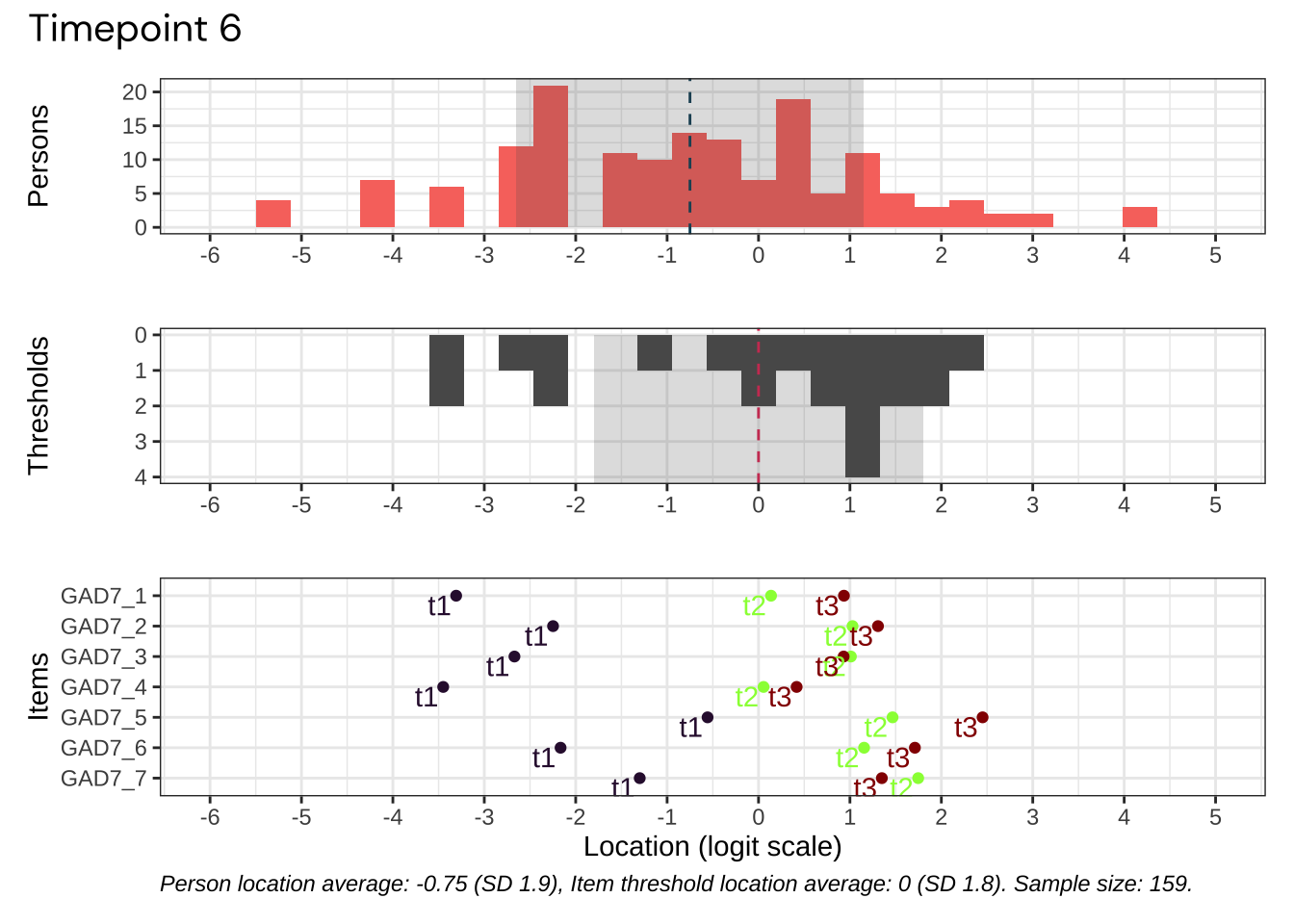
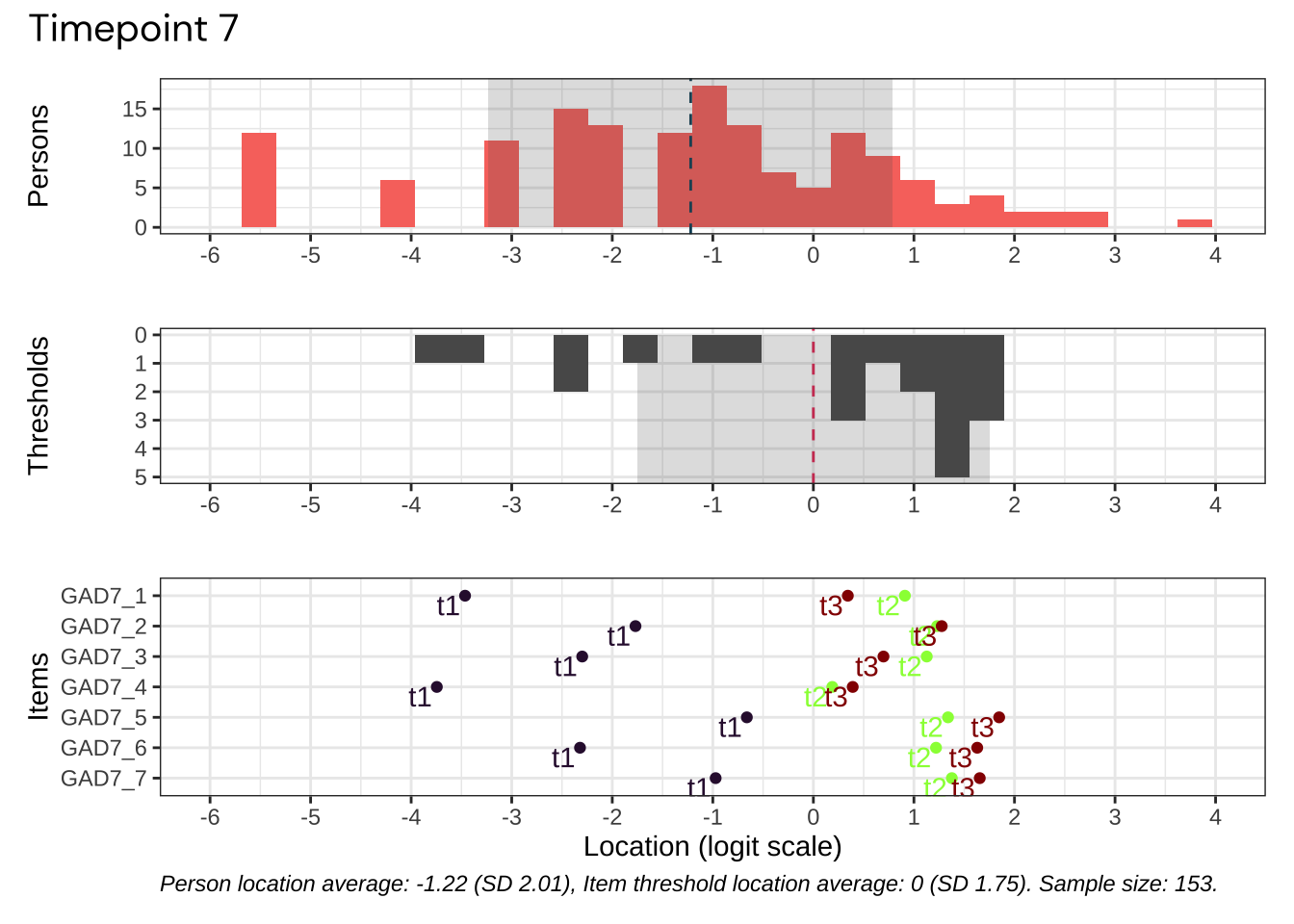
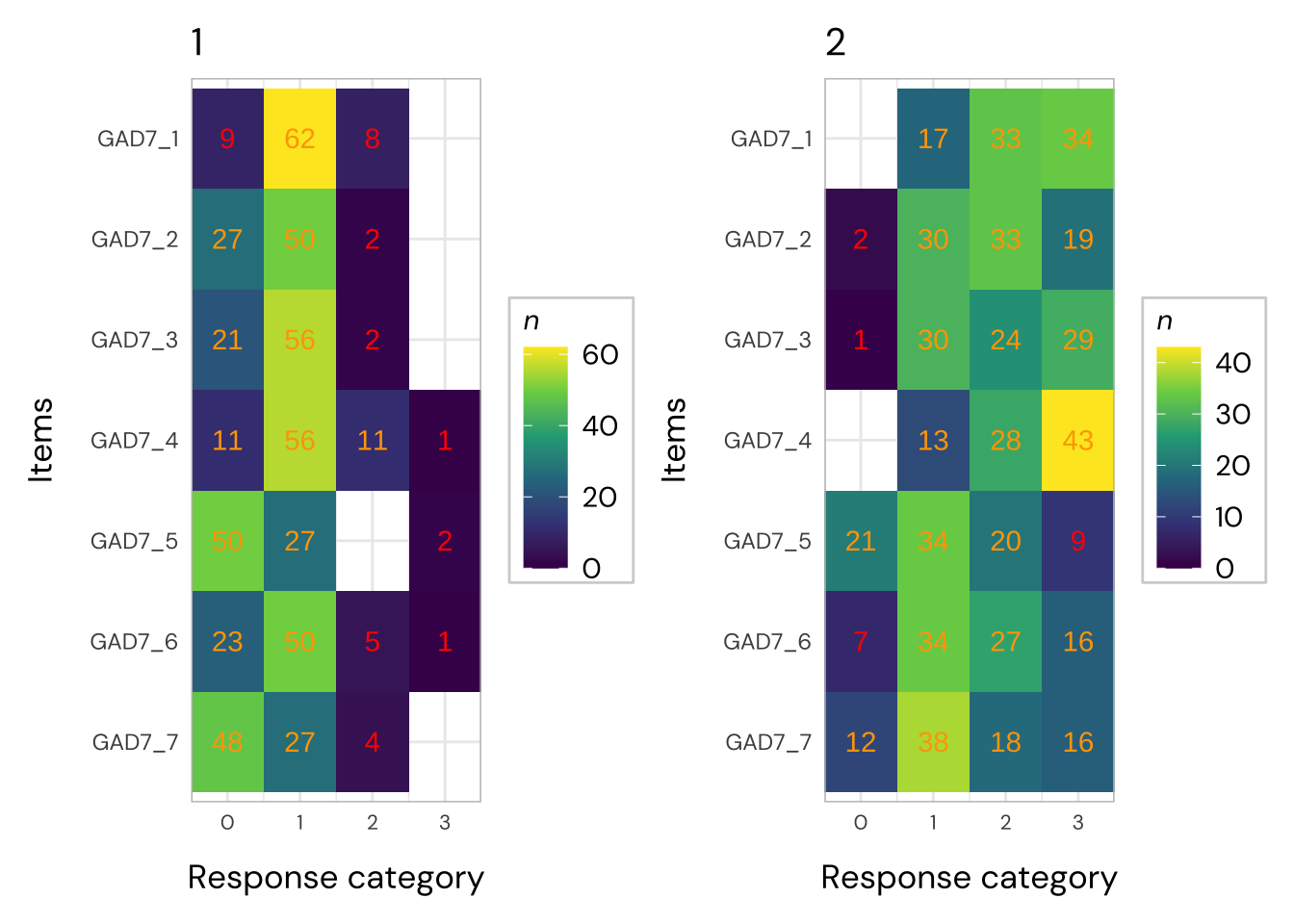
Code plots [[ 1 ] ] plots [[ 2 ] ] plots [[ 3 ] ] plots [[ 4 ] ] plots [[ 5 ] ] plots [[ 6 ] ] plots [[ 7 ] ] plots [[ 8 ] ] plots [[ 9 ] ] plots [[ 10 ] ] Looks like timepoint 2 is best targeted, and has a sample size of 163 complete responses.
Rasch analysis 1
The eRm package, which uses Conditional Maximum Likelihood (CML) estimation, will be used primarily. For this analysis, the Partial Credit Model will be used.
itemnr
item
GAD7_1
Känt dig nervös, ängslig eller väldigt stressad
GAD7_2
Inte kunnat sluta oroa dig eller kontrollera din oro
GAD7_3
Oroat dig för mycket för olika saker
GAD7_4
Haft svårt att slappna av
GAD7_5
Varit så rastlös att du har haft svårt att sitta still
GAD7_6
Blivit lätt irriterad eller retlig
GAD7_7
Känt dig rädd för att något hemskt skulle hända
Code
Item
InfitMSQ
Infit thresholds
OutfitMSQ
Outfit thresholds
Infit diff
Outfit diff
Relative location
GAD7_1
0.693
[0.735, 1.213]
0.642
[0.704, 1.42]
0.042
0.062
-0.61
GAD7_2
0.601
[0.834, 1.242]
0.584
[0.832, 1.26]
0.233
0.248
0.42
GAD7_3
0.651
[0.791, 1.314]
0.608
[0.758, 1.302]
0.14
0.15
0.06
GAD7_4
0.799
[0.774, 1.256]
0.758
[0.751, 1.359]
no misfit
no misfit
-0.71
GAD7_5
1.576
[0.773, 1.178]
1.94
[0.712, 1.475]
0.398
0.465
1.35
GAD7_6
1.281
[0.752, 1.396]
1.406
[0.74, 1.488]
no misfit
no misfit
0.52
GAD7_7
1.334
[0.811, 1.202]
1.328
[0.772, 1.534]
0.132
no misfit
1.02
Note:
Code
Item
Observed value
Model expected value
Absolute difference
Adjusted p-value (BH)
Statistical significance level
Location
Relative location
GAD7_1
0.77
0.64
0.13
0.001
***
-0.91
-0.61
GAD7_2
0.81
0.62
0.19
0.000
***
0.12
0.42
GAD7_3
0.81
0.65
0.16
0.000
***
-0.23
0.06
GAD7_4
0.73
0.64
0.09
0.072
.
-1.00
-0.71
GAD7_5
0.42
0.62
0.20
0.020
*
1.06
1.35
GAD7_6
0.51
0.62
0.11
0.098
.
0.23
0.52
GAD7_7
0.51
0.62
0.11
0.096
.
0.73
1.02
Code
PCA of Rasch model residuals
Eigenvalues
Proportion of variance
1.94
30.8%
1.48
23.4%
1.15
21.7%
0.90
10.2%
0.79
7.2%
Code
GAD7_1
GAD7_2
GAD7_3
GAD7_4
GAD7_5
GAD7_6
GAD7_7
GAD7_1
GAD7_2
0.27
GAD7_3
0.14
0.21
GAD7_4
0.16
0.04
-0.11
GAD7_5
-0.36
-0.32
-0.44
-0.07
GAD7_6
-0.26
-0.32
-0.13
-0.23
-0.08
GAD7_7
-0.28
-0.16
-0.06
-0.37
-0.18
-0.18
Note:
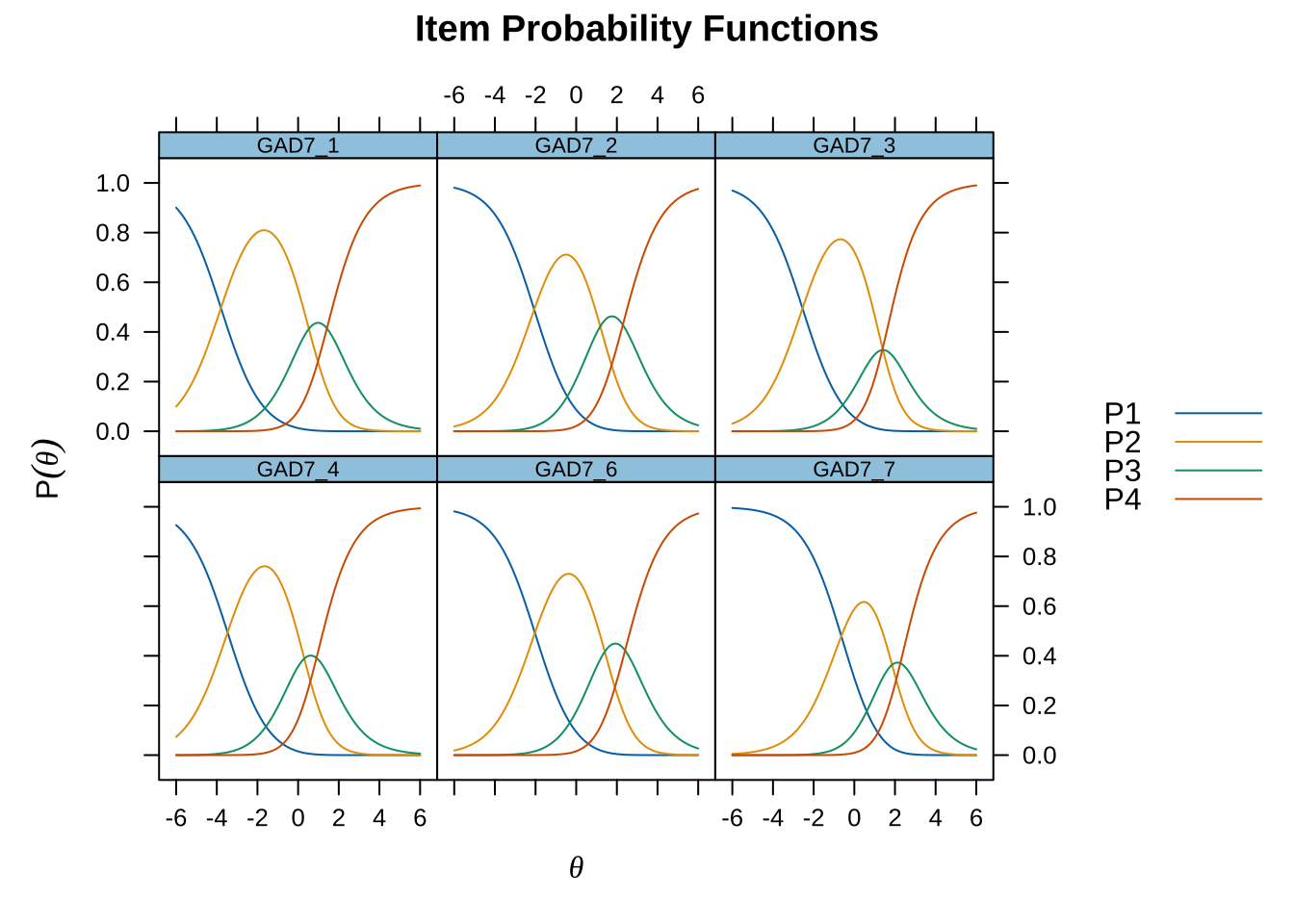
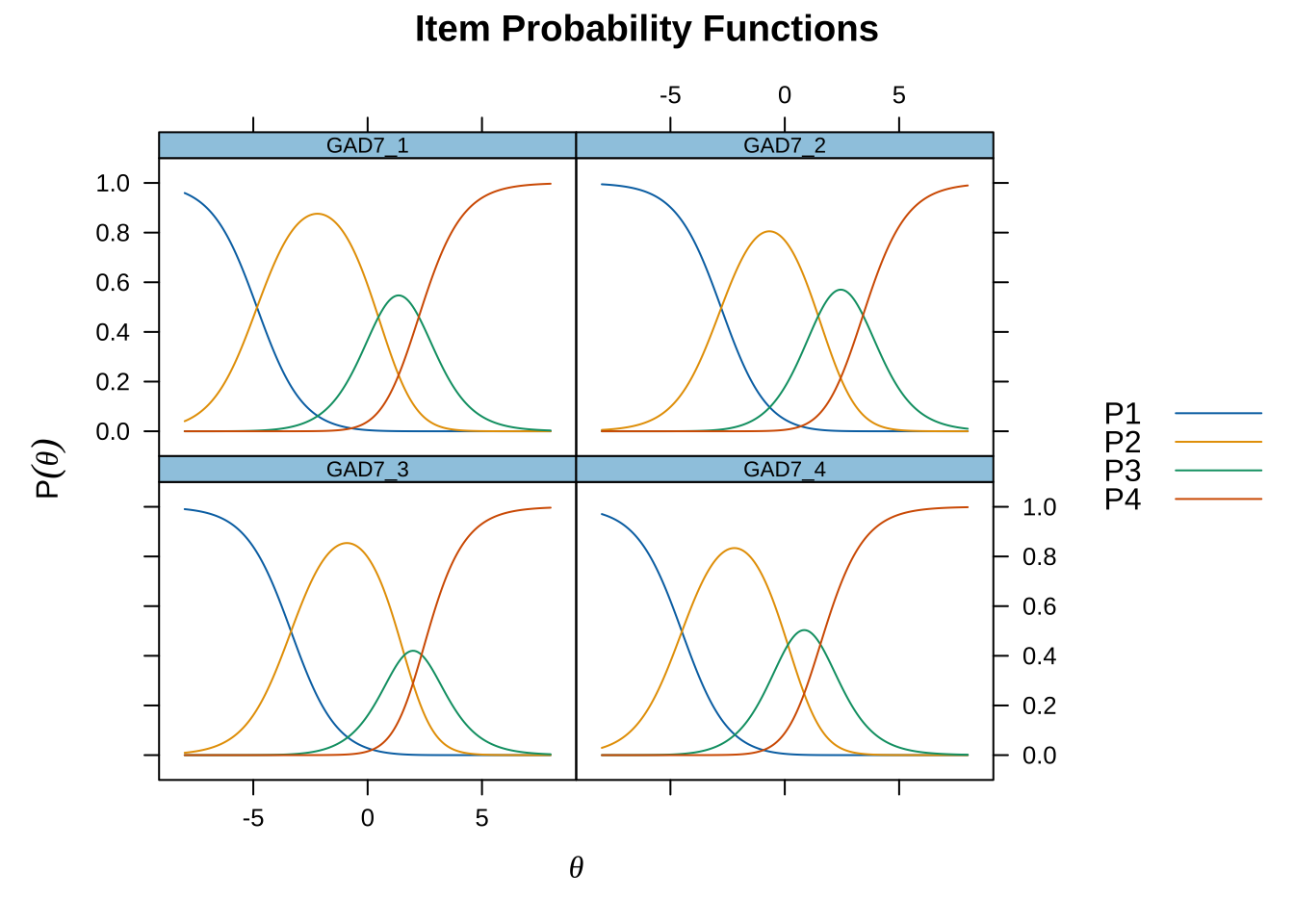
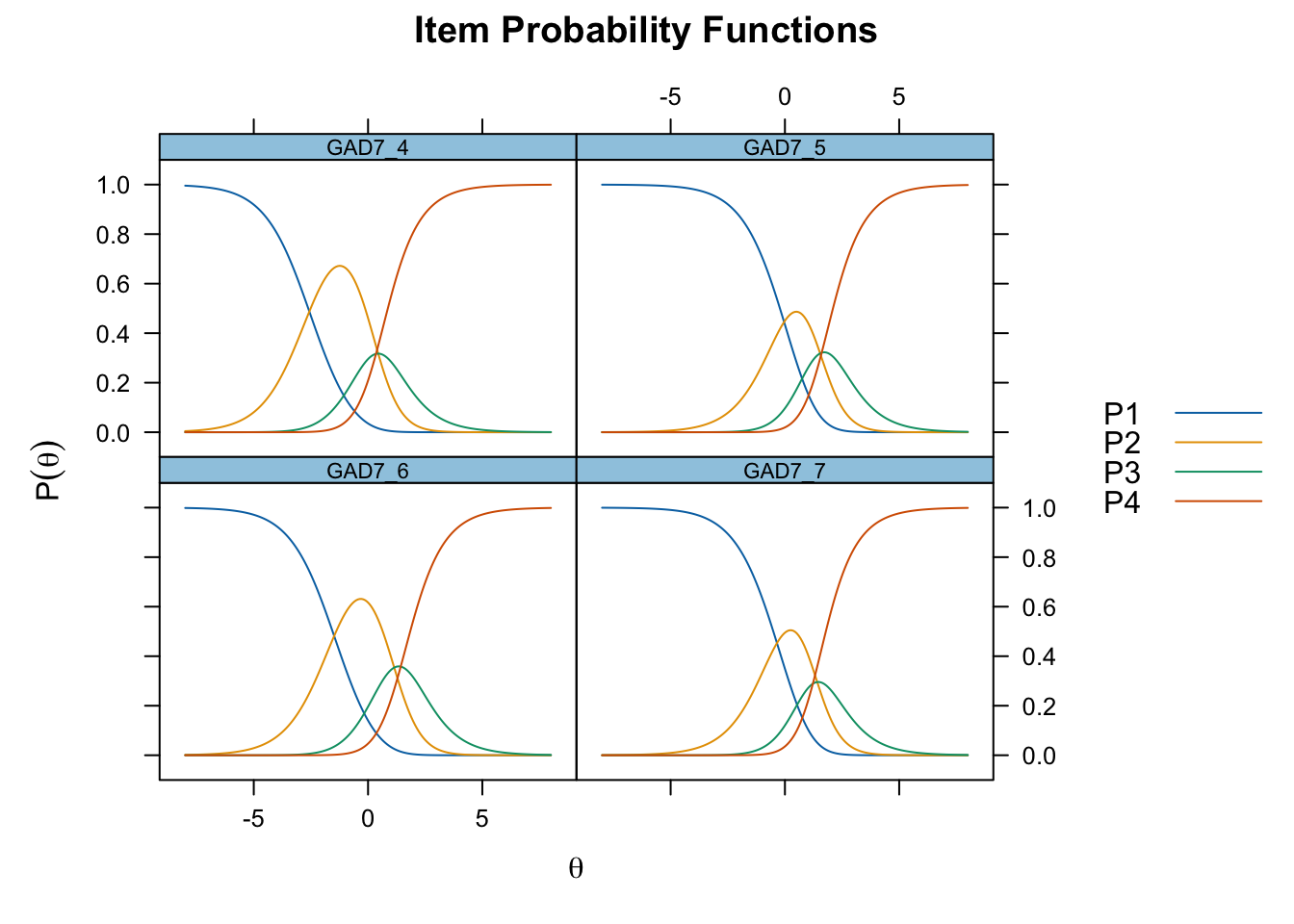
Code mirt ( d , model= 1 , itemtype= 'Rasch' , verbose = FALSE ) %>% plot ( type= "trace" , as.table = TRUE ,
theta_lim = c ( - 6 ,6 ) ) Code # for fewer items or a more magnified figure, use: #RIitemCats(df)
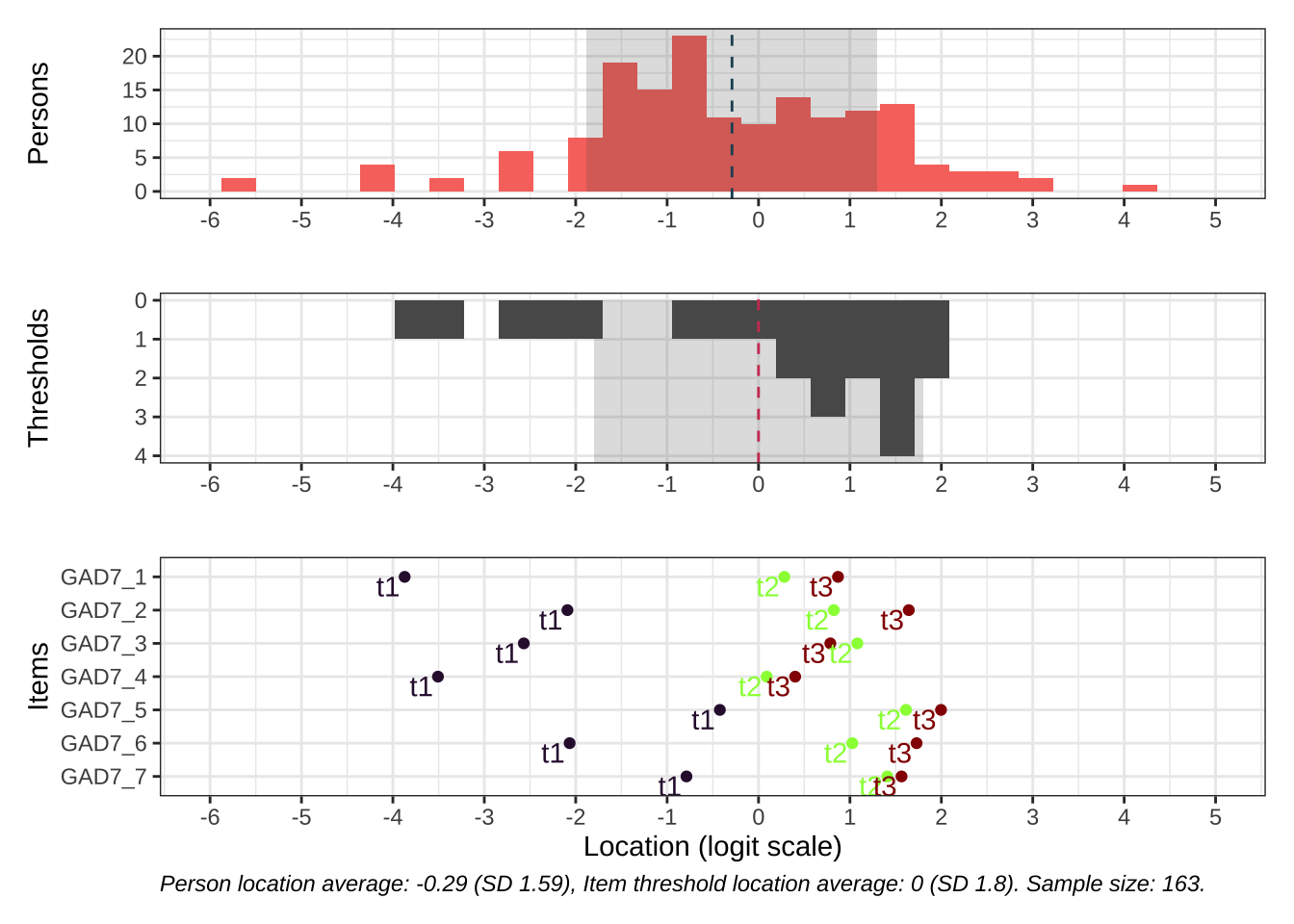
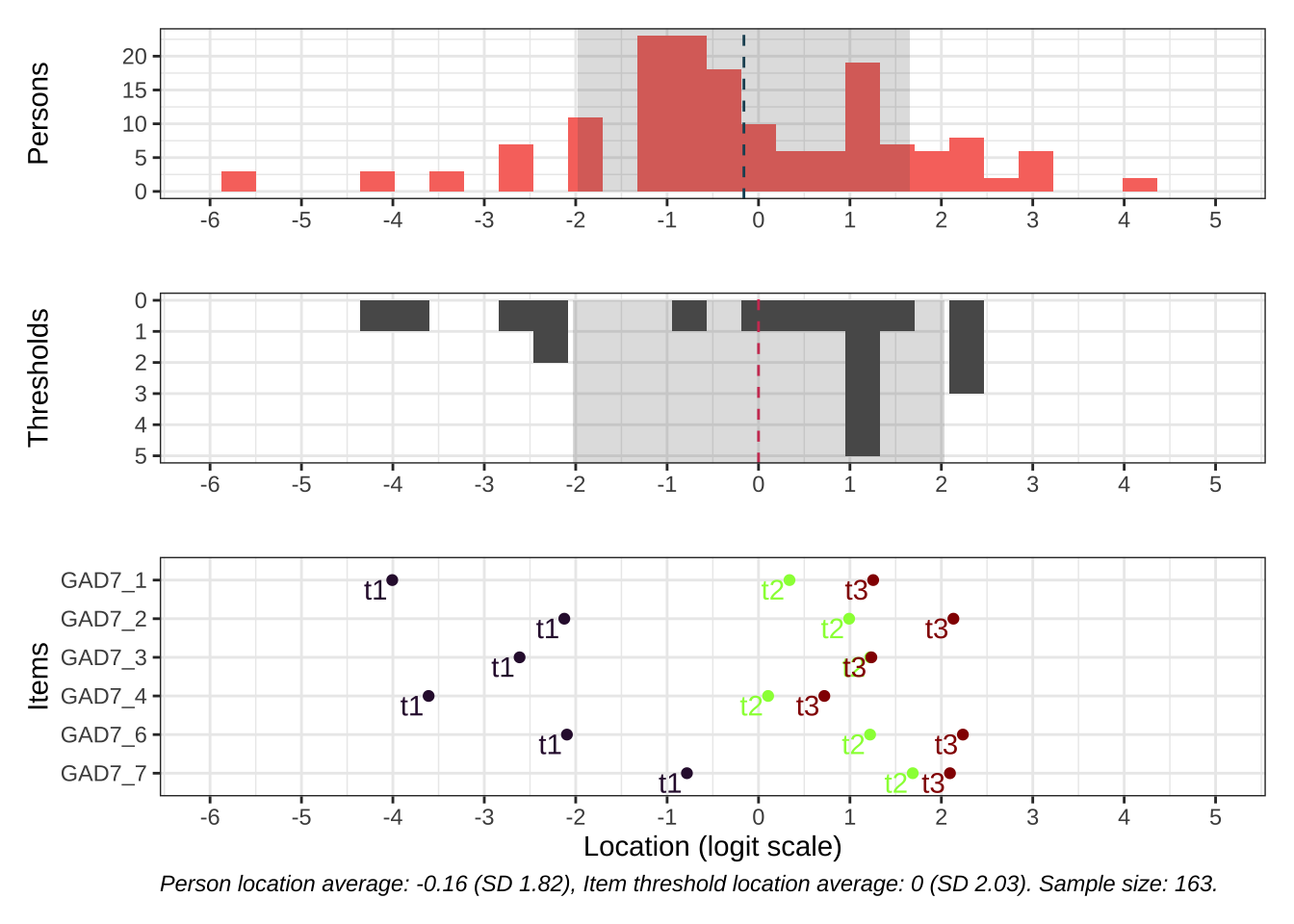
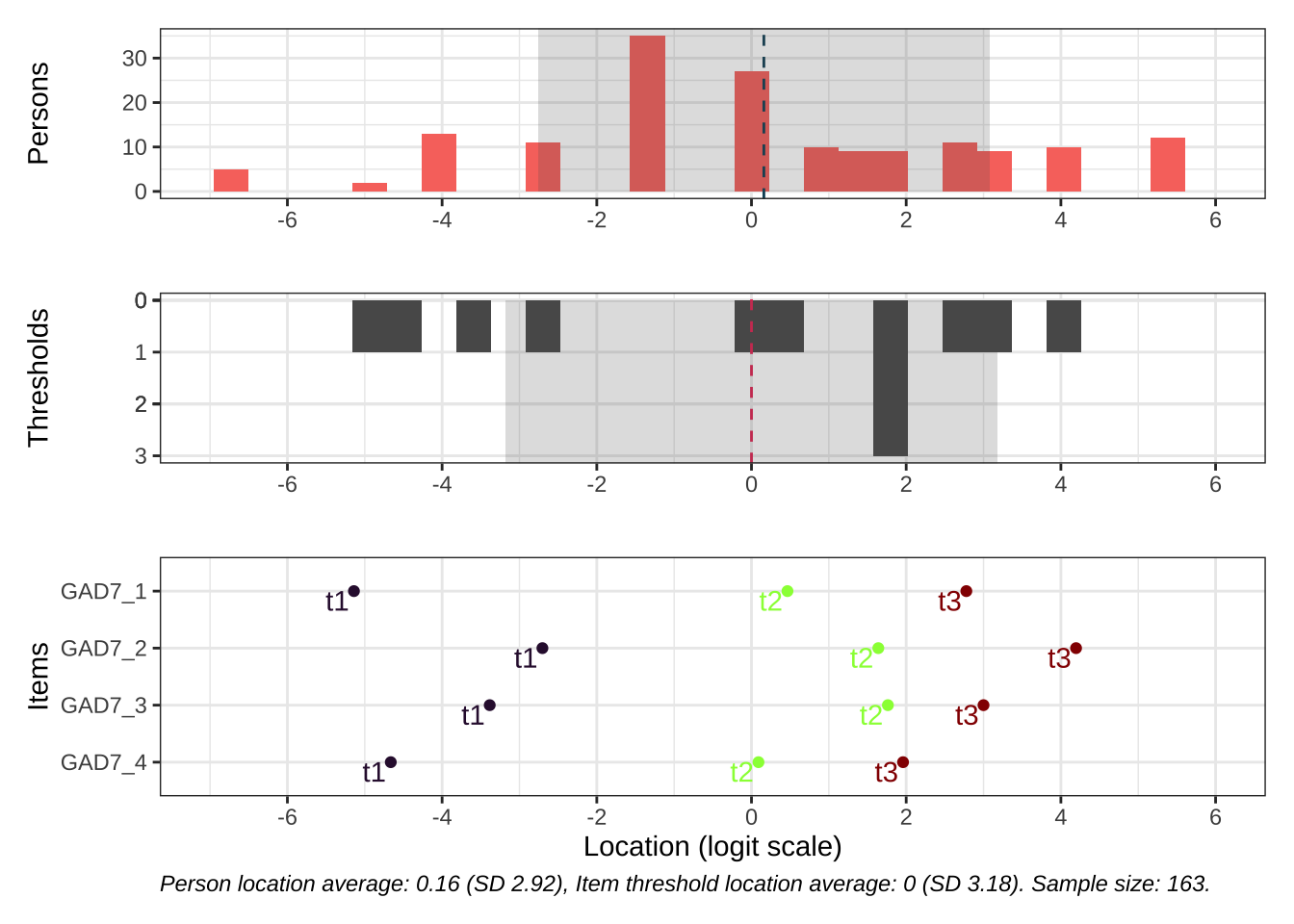
Code # increase fig-height above as needed, if you have many items RItargeting ( d )
Code
score_group n
1 1 79
2 2 84
Code
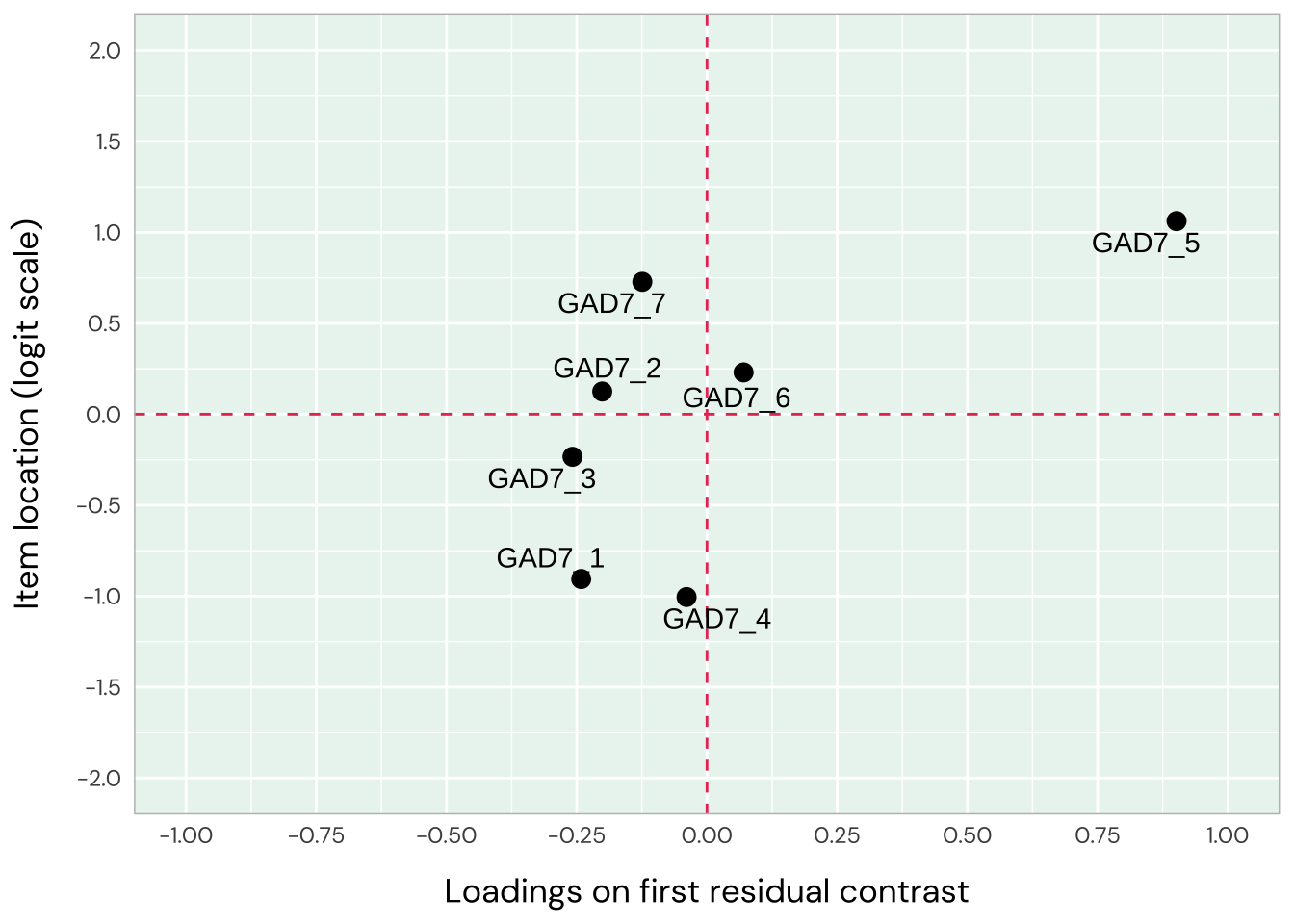
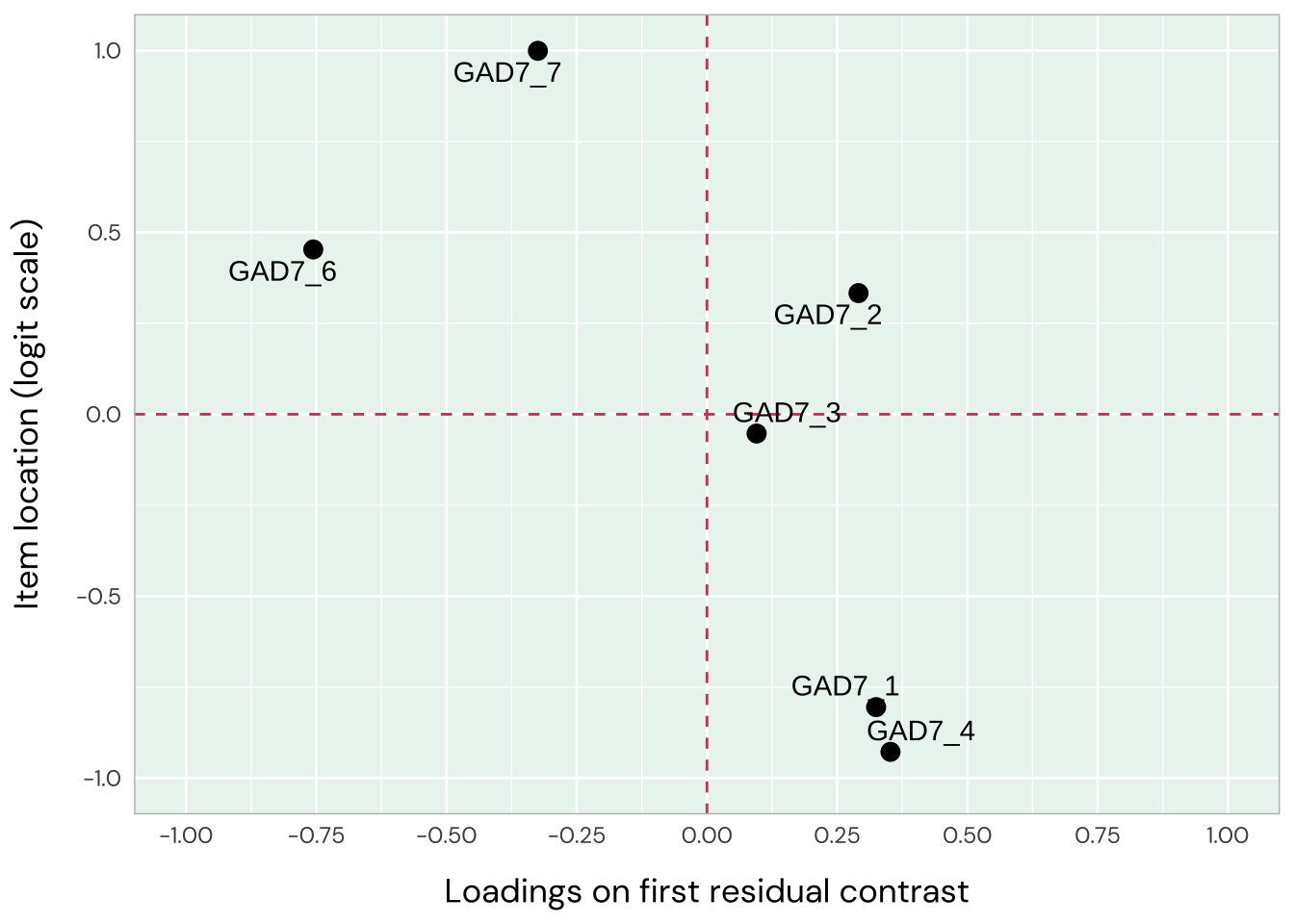
Item 1 has residual correlations with item 2 and 4, while 2 and 3 are also locally dependent. Items 1-3 are all overfit, whileiItems 5-7 are underfit (high item infit, particularly item 5 and to a lesser degree 7). This indicates two dimensions in data. The loadings plot does not show this as clearly, though, with item 5 being the only item deviating strongly.
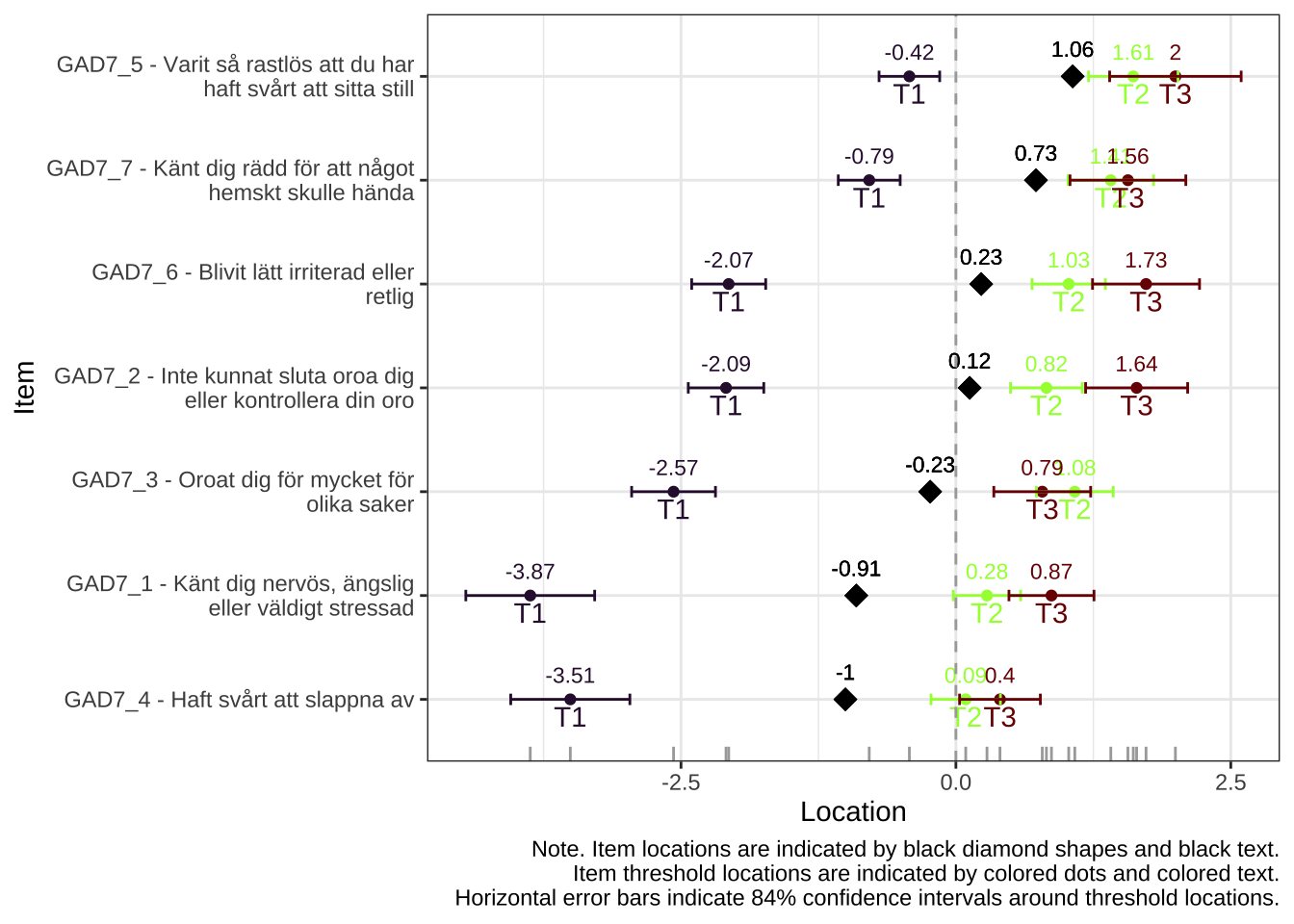
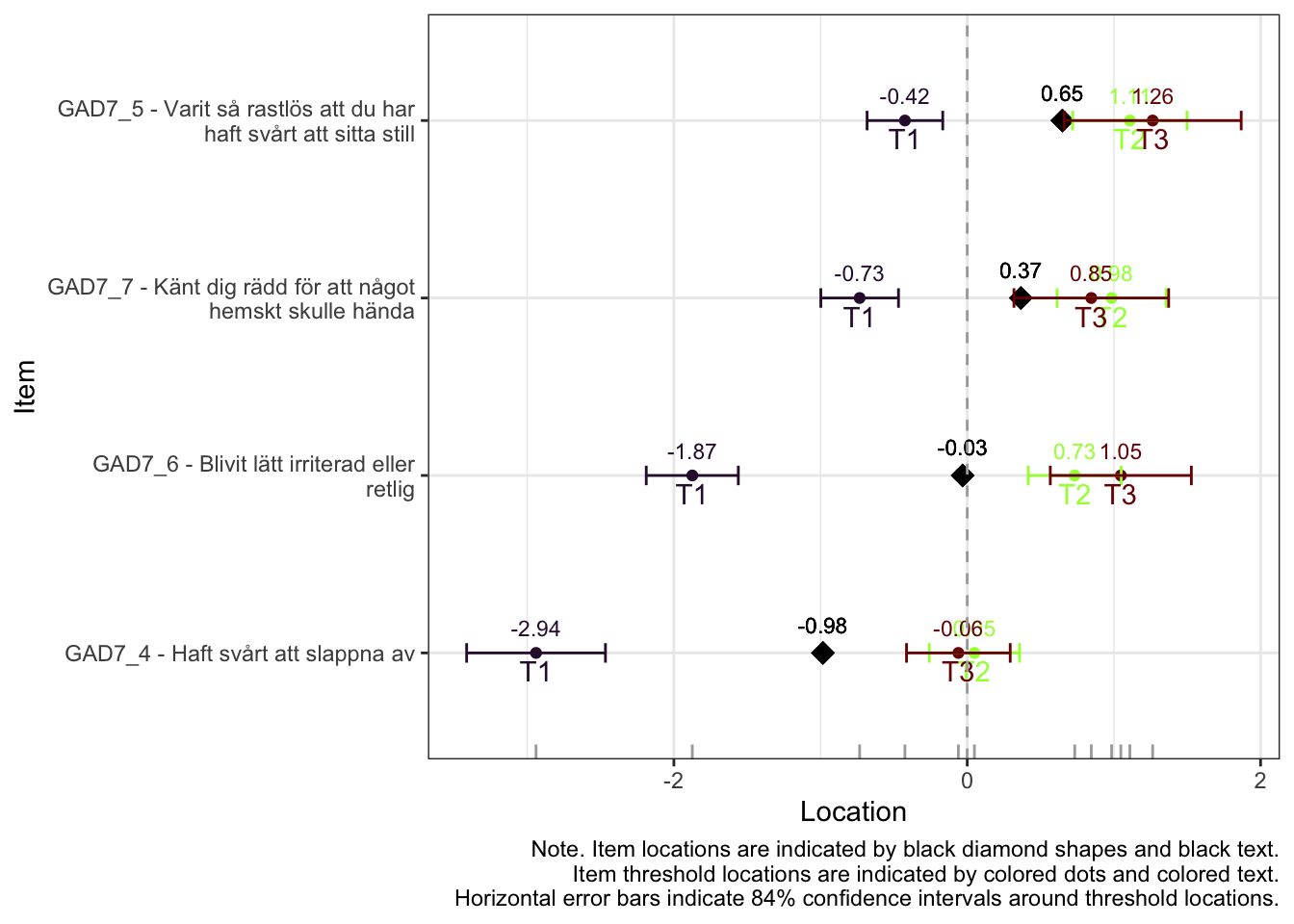
All items have issues with the second highest response category having very small distance from the adjancent item category threshold locations.
The sample is too small to create score groups for DIF analysis based on low/high scores.
Rasch analysis 2
While two dimensions seems likely, let’s see if we can find a working set of unidimensional items based on all 7. First, we remove item 5, leaving other issues for later.
Code d.backup <- d d $ GAD7_5 <- NULL
itemnr
item
GAD7_1
Känt dig nervös, ängslig eller väldigt stressad
GAD7_2
Inte kunnat sluta oroa dig eller kontrollera din oro
GAD7_3
Oroat dig för mycket för olika saker
GAD7_4
Haft svårt att slappna av
GAD7_6
Blivit lätt irriterad eller retlig
GAD7_7
Känt dig rädd för att något hemskt skulle hända
Code
Item
InfitMSQ
Infit thresholds
OutfitMSQ
Outfit thresholds
Infit diff
Outfit diff
Relative location
GAD7_1
0.718
[0.73, 1.312]
0.68
[0.732, 1.417]
0.012
0.052
-0.64
GAD7_2
0.612
[0.752, 1.275]
0.606
[0.741, 1.314]
0.14
0.135
0.50
GAD7_3
0.631
[0.74, 1.304]
0.574
[0.713, 1.365]
0.109
0.139
0.11
GAD7_4
0.975
[0.793, 1.256]
1.012
[0.725, 1.243]
no misfit
no misfit
-0.76
GAD7_6
1.501
[0.793, 1.22]
1.633
[0.764, 1.369]
0.281
0.264
0.62
GAD7_7
1.514
[0.738, 1.32]
1.452
[0.694, 1.628]
0.194
no misfit
1.16
Note:
Code
Item
Observed value
Model expected value
Absolute difference
Adjusted p-value (BH)
Statistical significance level
Location
Relative location
GAD7_1
0.81
0.69
0.12
0.001
***
-0.80
-0.64
GAD7_2
0.82
0.67
0.15
0.000
***
0.33
0.50
GAD7_3
0.85
0.70
0.15
0.000
***
-0.05
0.11
GAD7_4
0.71
0.69
0.02
0.721
-0.93
-0.76
GAD7_6
0.49
0.67
0.18
0.022
*
0.45
0.62
GAD7_7
0.52
0.66
0.14
0.037
*
1.00
1.16
Code
PCA of Rasch model residuals
Eigenvalues
Proportion of variance
1.81
33.7%
1.36
30.8%
1.10
16.1%
0.88
10.1%
0.84
9.1%
Code
GAD7_1
GAD7_2
GAD7_3
GAD7_4
GAD7_6
GAD7_7
GAD7_1
GAD7_2
0.18
GAD7_3
-0.01
0.08
GAD7_4
0.1
-0.01
-0.21
GAD7_6
-0.33
-0.39
-0.2
-0.23
GAD7_7
-0.37
-0.25
-0.15
-0.4
-0.19
Note:
Code mirt ( d , model= 1 , itemtype= 'Rasch' , verbose = FALSE ) %>% plot ( type= "trace" , as.table = TRUE ,
theta_lim = c ( - 6 ,6 ) )
Code # increase fig-height above as needed, if you have many items RItargeting ( d )
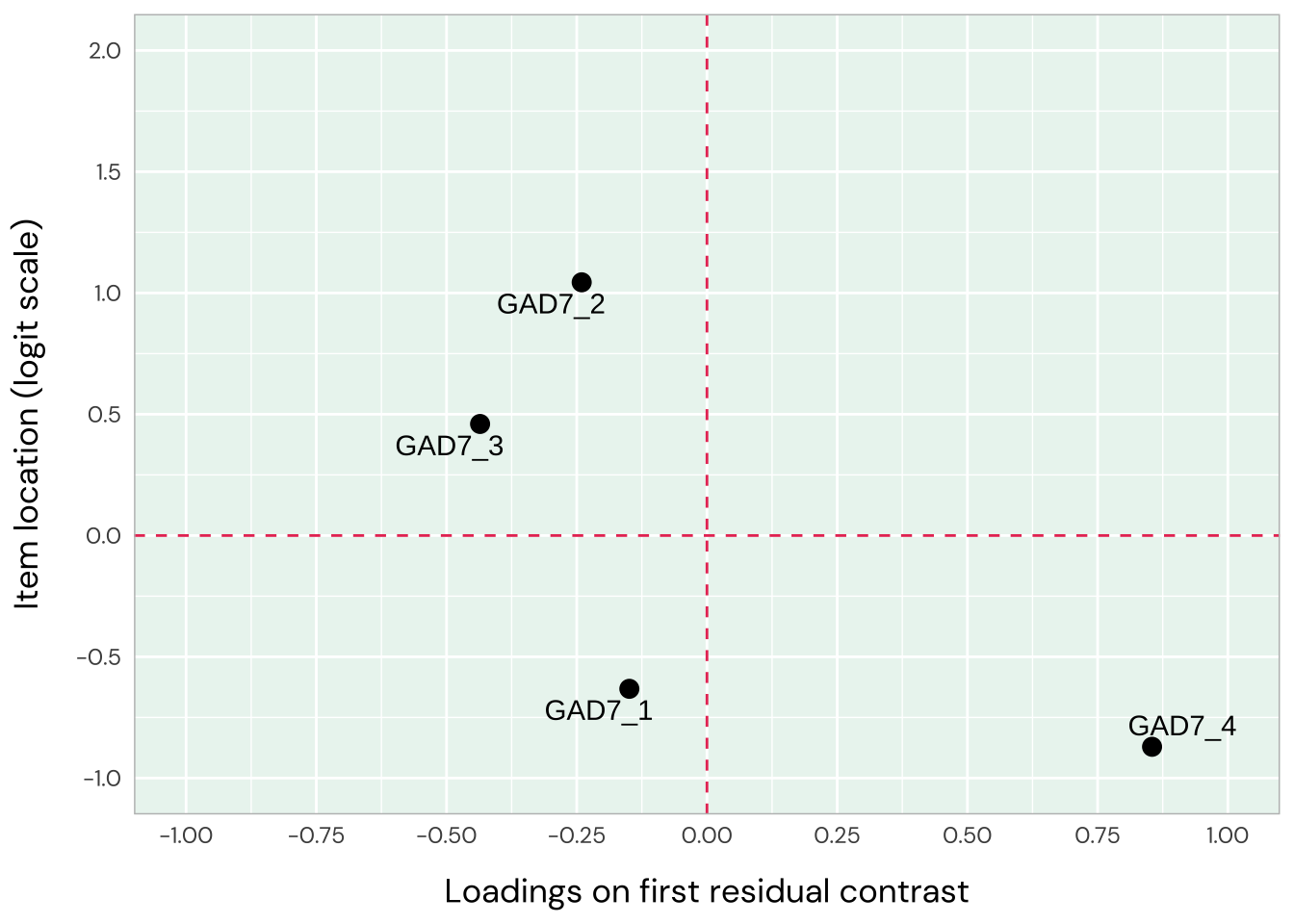
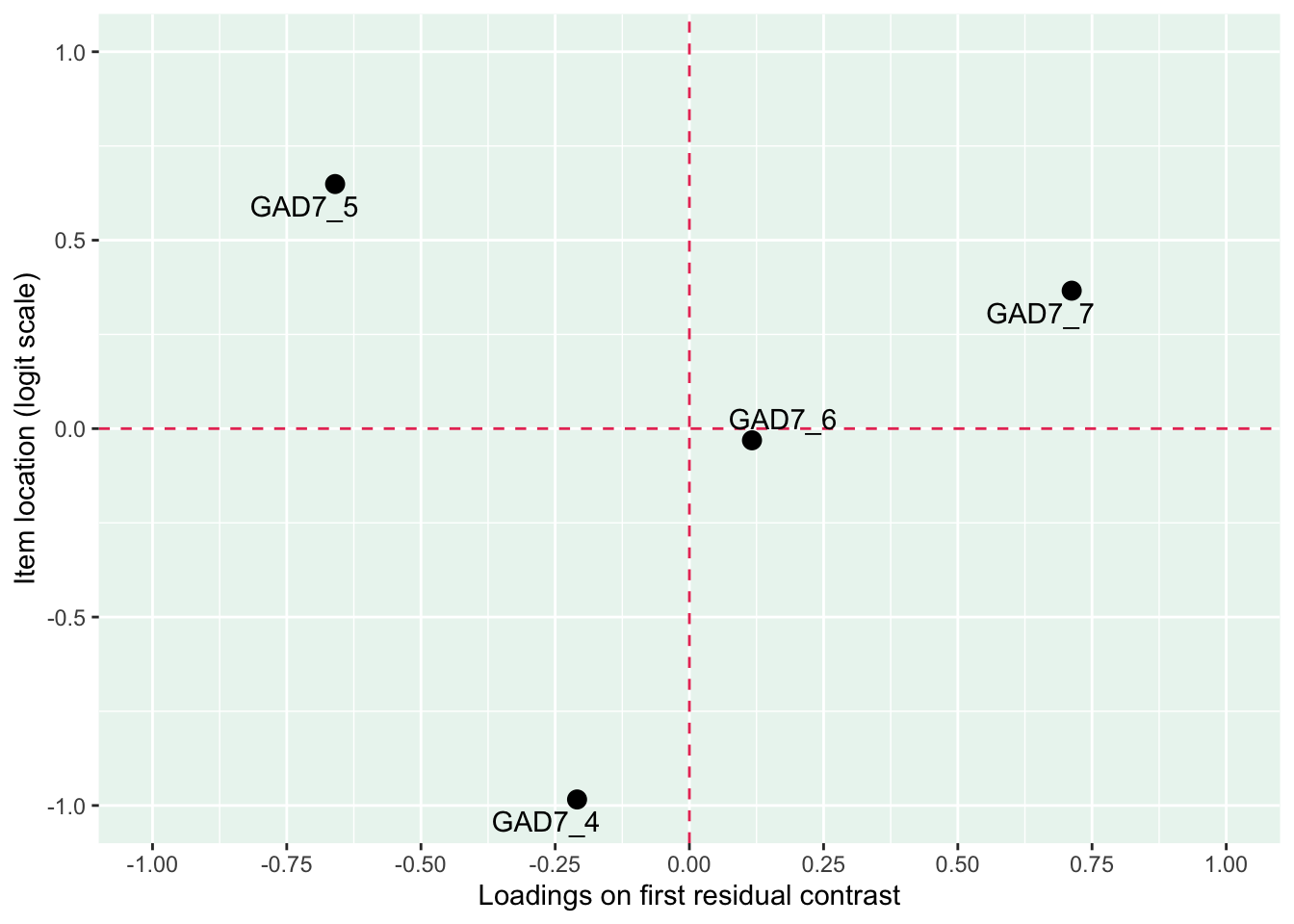
Only items 1 and 2 have a residual correlation now. The pattern in underfit and overfit items is more clearly seen in the loadings/location plot, with items 6 and 7 deviating. This confirms the two-dimensional structure, and we’ll move to look at items 1-4 and 4-7 separately.
Two separate dimensions
The initial analysis with all 7 items indicated that items 1-3 have low item fit (overfit to the Rasch model) strong residual correlations amongst them. This could mean that these items work well together, but likely represent another dimension than items 5-7, which in turn all showed high item fit (underfit).
We’ll make an analysis of this setup, including item 4 with both sets.
Code d1 <- d [ ,1 : 4 ] d2 <- d.backup [ ,4 : 7 ]
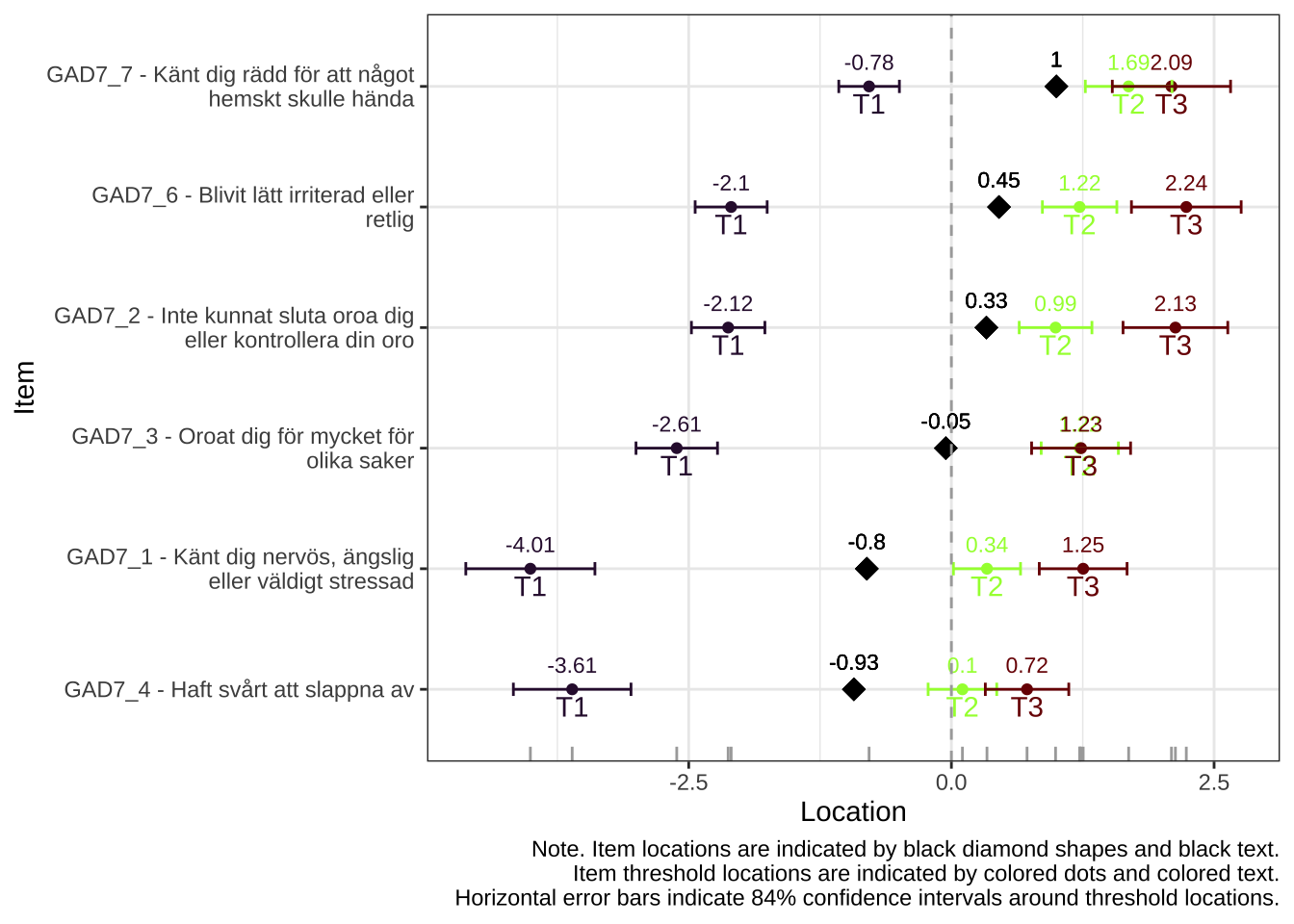
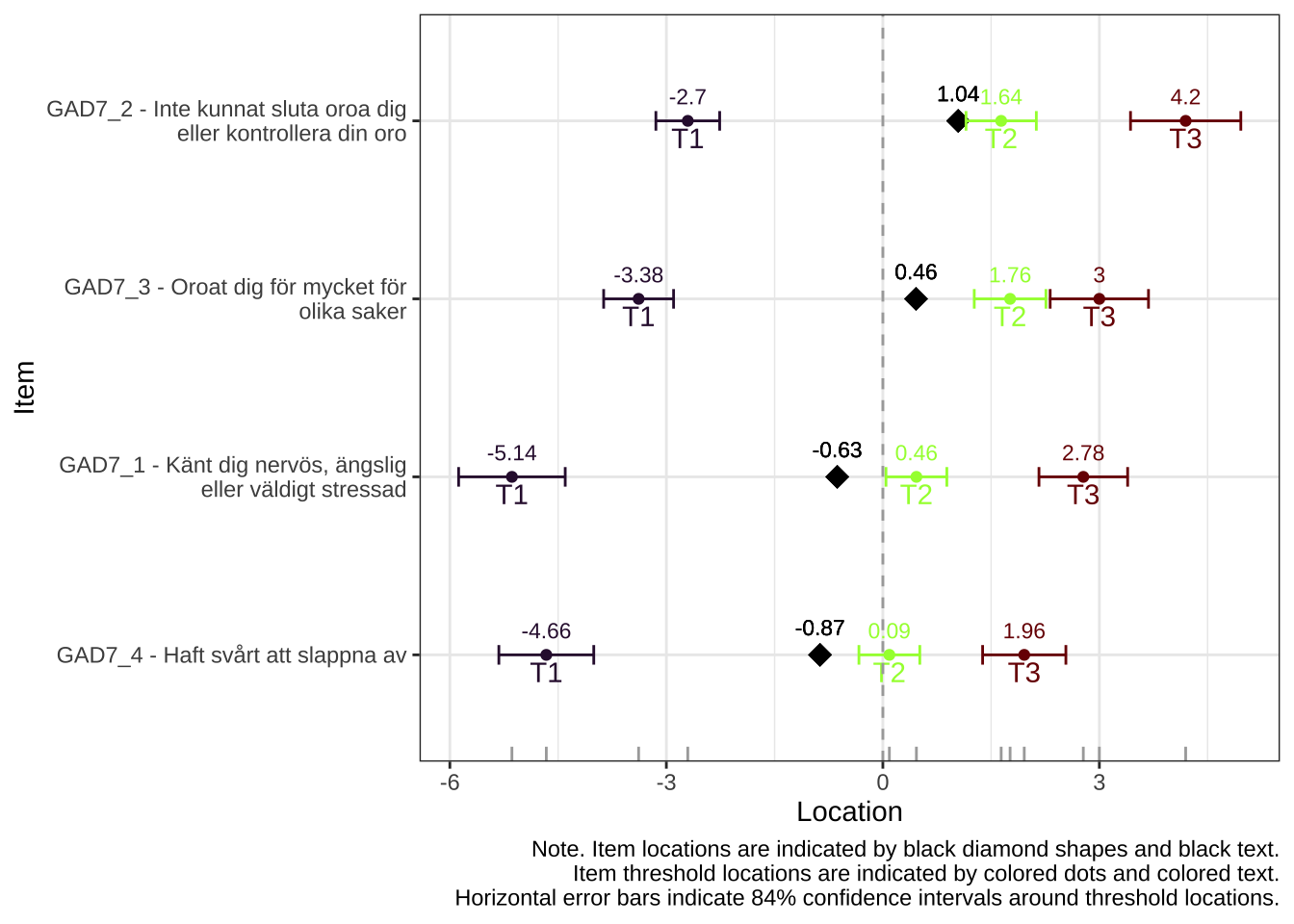
Items 1-4, worry
itemnr
item
GAD7_1
Känt dig nervös, ängslig eller väldigt stressad
GAD7_2
Inte kunnat sluta oroa dig eller kontrollera din oro
GAD7_3
Oroat dig för mycket för olika saker
GAD7_4
Haft svårt att slappna av
Code
Item
InfitMSQ
Infit thresholds
OutfitMSQ
Outfit thresholds
Infit diff
Outfit diff
Relative location
GAD7_1
0.845
[0.755, 1.251]
0.725
[0.696, 1.523]
no misfit
no misfit
-0.79
GAD7_2
0.781
[0.845, 1.25]
0.739
[0.742, 1.381]
0.064
0.003
0.88
GAD7_3
1.107
[0.686, 1.298]
0.891
[0.618, 1.735]
no misfit
no misfit
0.30
GAD7_4
1.264
[0.779, 1.255]
1.314
[0.609, 1.36]
0.009
no misfit
-1.03
Note:
Code
Item
Observed value
Model expected value
Absolute difference
Adjusted p-value (BH)
Statistical significance level
Location
Relative location
GAD7_1
0.88
0.83
0.05
0.177
-0.63
-0.79
GAD7_2
0.88
0.83
0.05
0.214
1.04
0.88
GAD7_3
0.84
0.85
0.01
0.824
0.46
0.30
GAD7_4
0.79
0.82
0.03
0.577
-0.87
-1.03
Code simpca <- RIbootPCA ( d ,200 , cpu = 8 ) simpca $ max Code
PCA of Rasch model residuals
Eigenvalues
Proportion of variance
1.49
44.8%
1.29
29.7%
1.20
25.1%
0.02
0.4%
Code
GAD7_1
GAD7_2
GAD7_3
GAD7_4
GAD7_1
GAD7_2
-0.14
GAD7_3
-0.27
-0.14
GAD7_4
-0.24
-0.33
-0.45
Note:
Code mirt ( d , model= 1 , itemtype= 'Rasch' , verbose = FALSE ) %>% plot ( type= "trace" , as.table = TRUE ,
theta_lim = c ( - 8 ,8 ) )
Code # increase fig-height above as needed, if you have many items RItargeting ( d )
Item-restscore looks good. Item infit shows slight overfit item 2. No residual correlations.
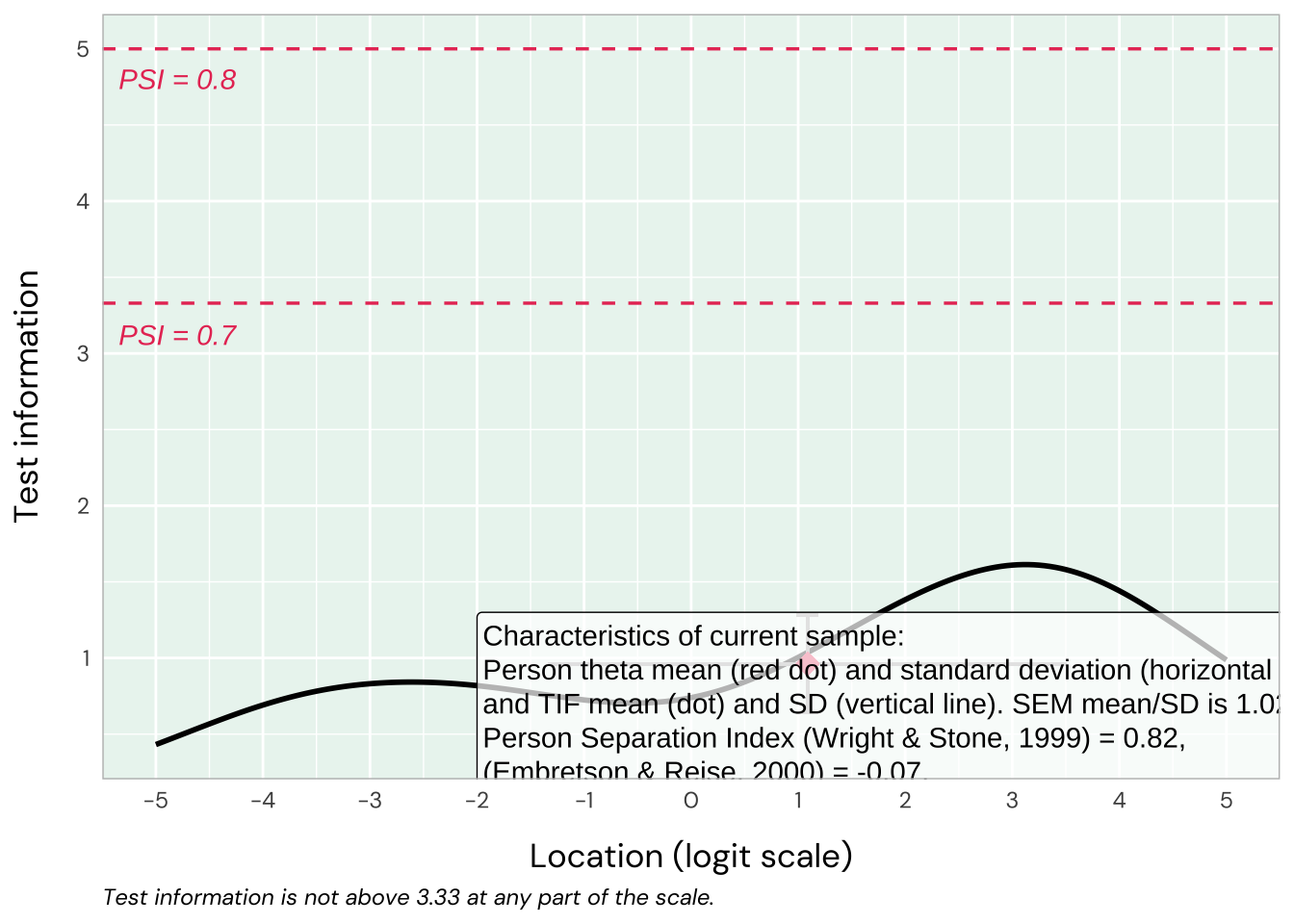
Reliability
TIF is low while PSI is ok at 0.82.
DIF
Code
[1] "No statistically significant DIF found."
Code
Item
2
3
Mean location
StDev
MaxDiff
GAD7_1
-0.693
-0.388
-0.540
0.216
0.306
GAD7_2
0.851
1.129
0.990
0.196
0.278
GAD7_3
0.304
0.256
0.280
0.034
0.048
GAD7_4
-0.461
-0.997
-0.729
0.379
0.536
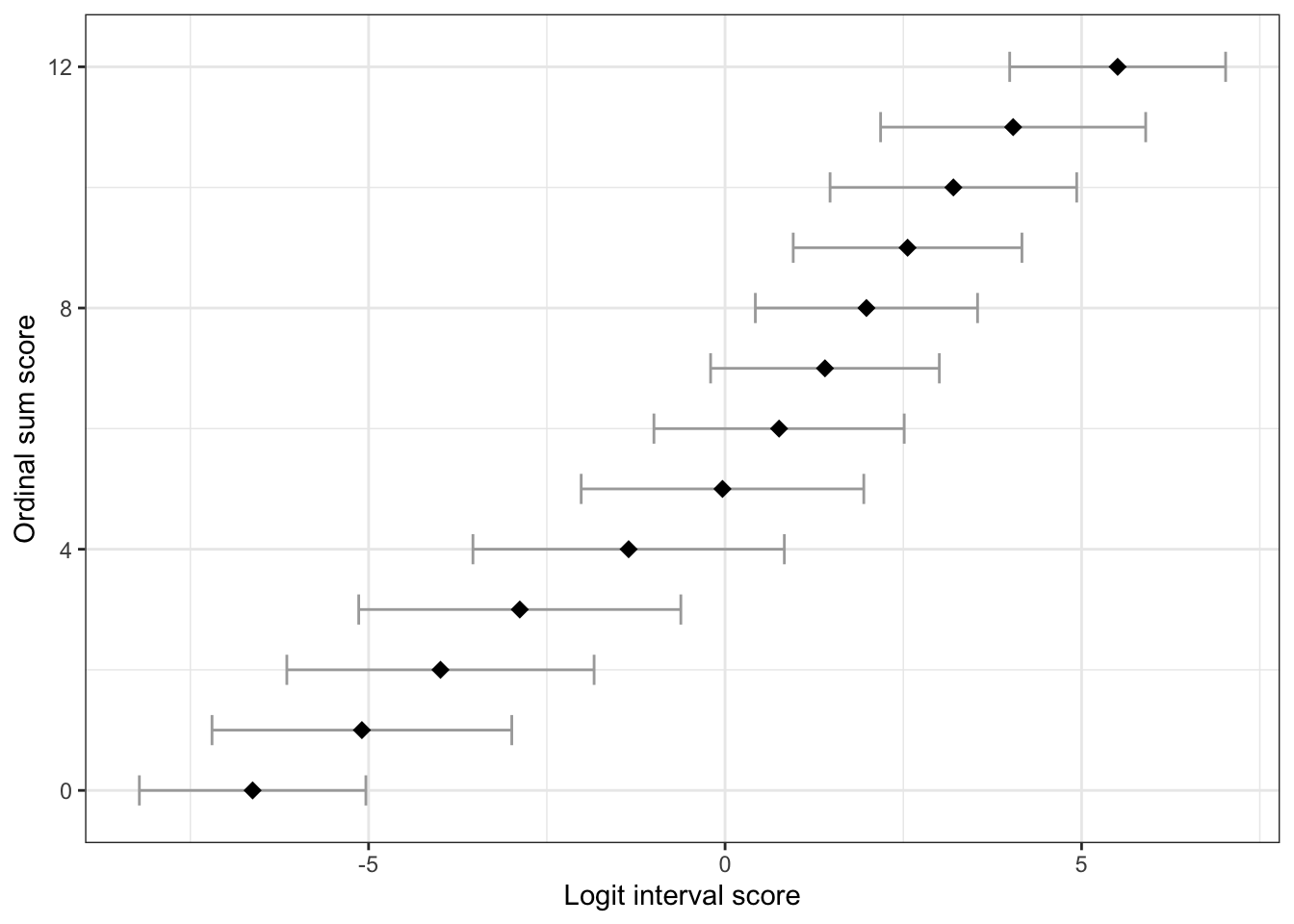
Item parameters
Code
Ordinal sum score
Logit score
Logit std.error
0
-6.626
0.810
1
-5.094
1.073
2
-3.992
1.099
3
-2.879
1.152
4
-1.353
1.114
5
-0.036
1.011
6
0.757
0.896
7
1.401
0.818
8
1.983
0.795
9
2.560
0.818
10
3.202
0.882
11
4.040
0.948
12
5.506
0.773
Code
Latent scores items 1-4
Code
Code
Min. 1st Qu. Median Mean 3rd Qu. Max.
-6.6262 -2.8791 -0.0363 -0.2278 1.9828 5.5062
Code
Items 4-7, anxiety
itemnr
item
GAD7_4
Haft svårt att slappna av
GAD7_5
Varit så rastlös att du har haft svårt att sitta still
GAD7_6
Blivit lätt irriterad eller retlig
GAD7_7
Känt dig rädd för att något hemskt skulle hända
Code
Item
InfitMSQ
Infit thresholds
OutfitMSQ
Outfit thresholds
Infit diff
Outfit diff
Relative location
GAD7_4
0.816
[0.833, 1.206]
0.788
[0.797, 1.382]
0.017
0.009
-0.59
GAD7_5
1.054
[0.817, 1.245]
1.034
[0.76, 1.487]
no misfit
no misfit
1.04
GAD7_6
0.982
[0.803, 1.166]
0.995
[0.819, 1.214]
no misfit
no misfit
0.36
GAD7_7
1.148
[0.793, 1.265]
1.055
[0.788, 1.253]
no misfit
no misfit
0.76
Note:
Code
Item
Observed value
Model expected value
Absolute difference
Adjusted p-value (BH)
Statistical significance level
Location
Relative location
GAD7_4
0.57
0.47
0.10
0.366
-0.98
-0.59
GAD7_5
0.43
0.46
0.03
0.760
0.65
1.04
GAD7_6
0.49
0.46
0.03
0.760
-0.03
0.36
GAD7_7
0.42
0.47
0.05
0.760
0.37
0.76
Code simpca <- RIbootPCA ( d ,200 , cpu = 8 ) simpca $ max Code
PCA of Rasch model residuals
Eigenvalues
Proportion of variance
1.45
37.7%
1.32
34.4%
1.23
27.6%
0.01
0.3%
Code
GAD7_4
GAD7_5
GAD7_6
GAD7_7
GAD7_4
GAD7_5
-0.11
GAD7_6
-0.15
-0.24
GAD7_7
-0.24
-0.34
-0.22
Note:
Code mirt ( d , model= 1 , itemtype= 'Rasch' , verbose = FALSE ) %>% plot ( type= "trace" , as.table = TRUE ,
theta_lim = c ( - 8 ,8 ) )
Code # increase fig-height above as needed, if you have many items RItargeting ( d )
Item 4 actually works quite well with items 5-7 too. However, it shows disordered thresholds in this item set, which it does not with items 1-3, so we will use it as a part of item set 1, “worry”.
Item thresholds have small distances or are disordered (2 items) in this item set, which is further indicated by the low reliability, even before merging response categories. We will not use this second set of items in any analysis.